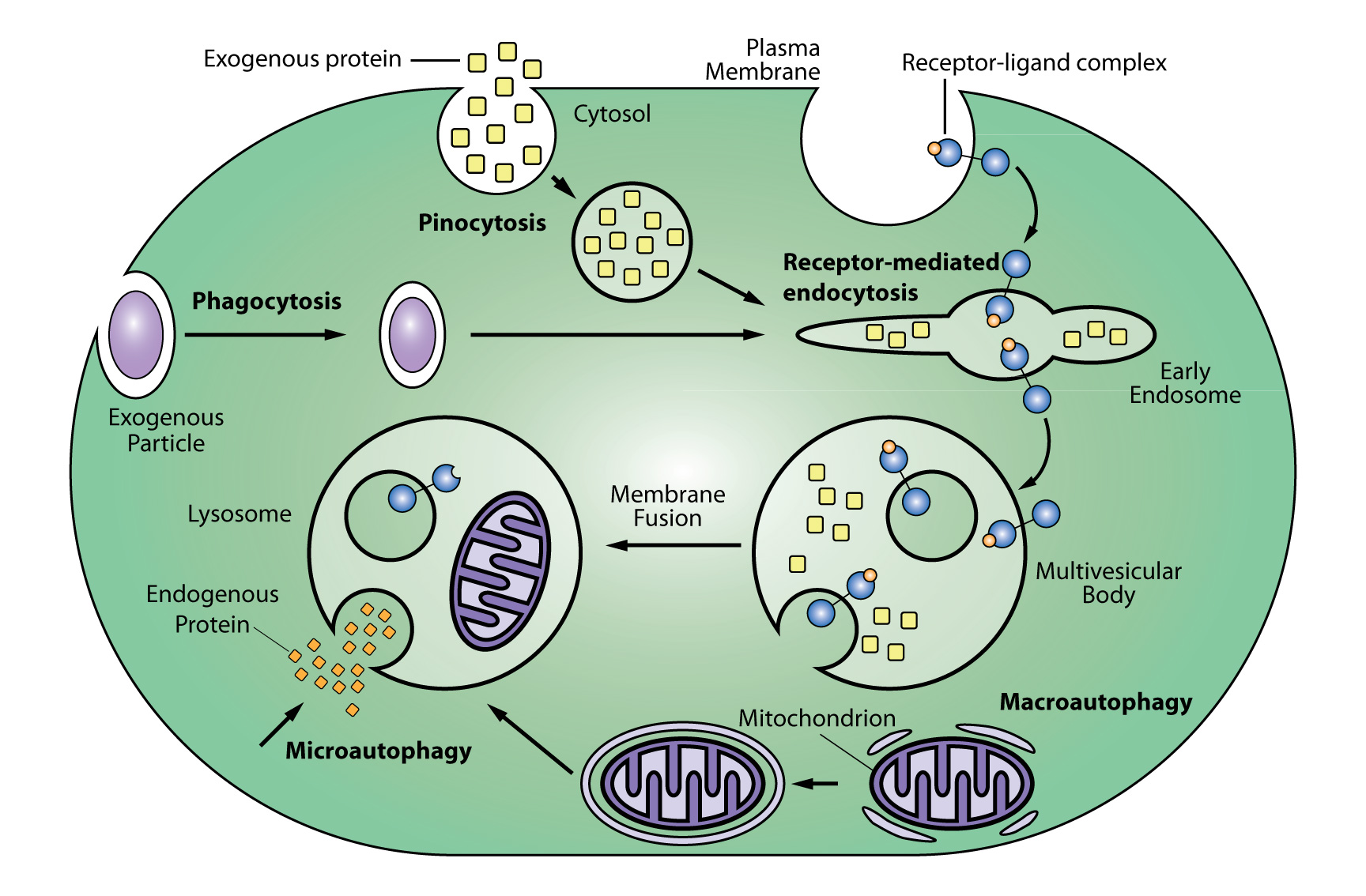
Endosomes and the lyosgenic pathway
Lysosomal proteolysis is one of two protein degradation pathways in cells (the other being the ubiquitin-proteasome system). Lysosomes are produced by the Golgi apparatus (ie, trans-Golgi network)and degrade extracellular proteins and molecules as well as cytoplasmic material and organelles (eg, mitochondria). Extracellular molecules enter a cell through endocytosis, phagocytosis, and pinocytosis while degradation of intracellular proteins and molecules involves microautophagy and macroautophagy. Following entry, extracellular molecules undergo processing and sorting by early endosomes, and molecules destined for degradation are passed onto late endosomes (aka, multivesicular bodies [MSVs]), and then membrane fusion of the late endosome with a lysosome that is a hybrid of the two structures. The journey of molecules destined for lysosomal degradation and/or the subcellular components of this pathway can be tracked using SiR-Lysosome in live cells (see below). Early endosomes are identified by expression of the small GTPases RAB5 and RAB4, as well as transferrin and its receptor and early endosome antigen 1 (EEA1). Late endosomes are identified by expression of the small GTPases RAB7 and RAB9 and mannose 6-phosphate receptors. A prototypical protein that is degraded by the lysosomes is epidermal growth factor (EGF) and the EGF receptor (EGFR).
Follow live cell invagination of endosomes and the lyosgenic process by using SiR-Lysosome red fluorecent dye or SiR700-Lysosome far-red fluorescent dye.
.
Endosome processing in the lysogen pathway.
Observe endosome / lysosome movement in live cells
Live cell video of mitotic cells stained with far-red SiR-Lysosome (yellow) and red SiR-Tubulin (red) fluorescent probes.
Image endosomes and lysosomes during cell motility
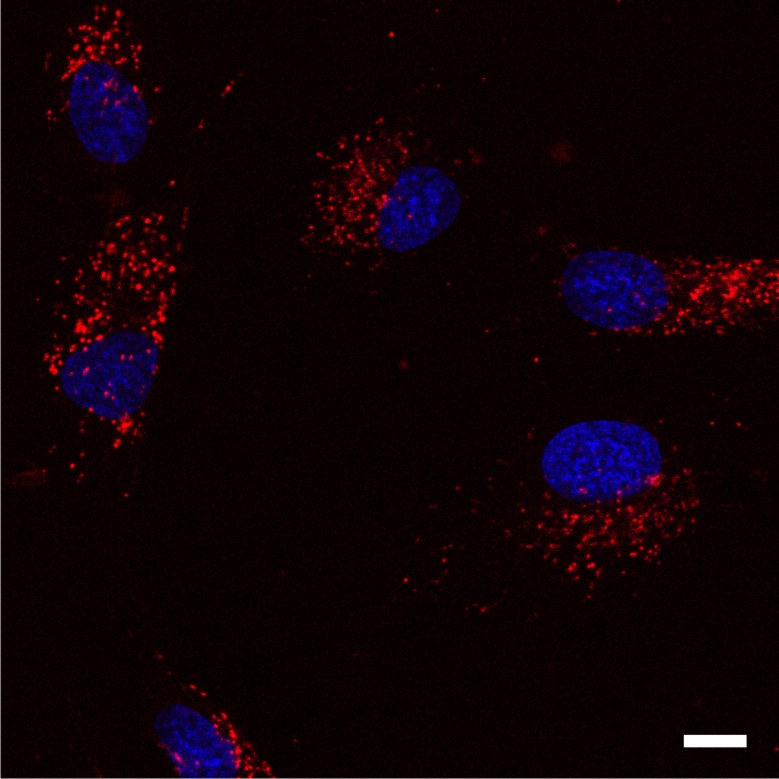
Live human fibroblast cells stained with 1 µM SiR700-Lysosome (red) and Hoechst (blue) for 1 h at 37°C and imaged by widefield microscopy.