Small G-protein News
The Ras superfamily of small GTPases consist of more than 150 members, which based on their sequence homology, are divided into several subfamilies such as Rho, Ras, Ran, Rab, Arf and Rad/Rem/Gem/Kir. This group of small GTPases serve as binary switches cycling between GDP-bound inactive and GTP-bound active state (1,2). The regulatory proteins for this switch include guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs) (1,2).
The Rho subfamily
The Rho family of small GTPases consists of at least 20 members, the most extensively characterized of which are the Rac1, RhoA and Cdc42 proteins (3). In common with all other small GTPases, the Rho proteins act as molecular switches that transmit cellular signals through an array of effector proteins. This family is involved in a wide range of cellular responses, including cytoskeletal reorganization (4-5), regulation of transcription (6), cell migration (7), cellular transformation and metastasis (8).
Click the links below to learn more about
Cdc42 Rac1 Rac2 Rac3 RhoA RhoB RhoC
The Ras subfamily
There are approximately 10 members of this subfamily split between Ras, Ral and Rap proteins. Ras proteins function in the Raf/ERK signaling pathway which controls proliferation and differentiation. Ral proteins function in the early steps of endocytosis, whereas Rap’s function as antagonist of Ras and they are located to the late endosomal and early lysosomal compartments.
Click the links below to learn more about
The Ran subfamily
The Ran proteins regulate nuclear transport of macromolecules, and in the cell cycle checkpoint from DNA replication to entry into mitosis.
The Rab subfamily
One of the largest subfamilies of small G-proteins, the Rab proteins regulate the flow of vesicles from the ER to the Golgi and onto the plasma membrane. They are ubiquitously expressed and are highly abundant.
Click here to learn more about Lysosomes
The Rad/Rem/Gem/Kir subfamily
Members of this subfamily (RGK subfamily) have wide ranging functions such as controlling cardiac hypertrophy, inhibiting insulin-stimulated glucose uptake in myocyte and adipocyte cell lines, as well as inhibiting voltage dependent calcium channels by binding to the b-subunit (9).
The Arf subfamily
The Arf family consists of Arf proteins and Arf-like proteins (Arl’s) (10). As a general rule the Arf proteins help regulate vesicle trafficking and membrane fusion in functions such as nuclear membrane fusion, negative regulation of Golgi vesicle transport, and plasma membrane invagination. This family is the most divergent of the small G-proteins being equally related to heterotrimeric G-protein family.
Small G-protein measurement
There are several methods used to measure Small G-proteins, these include a) measurement of the active "GTP-bound" form in extracts and in situ, b) measuring total Small G-protein levels, c) Exchange activity from GTP to GDP and vice versa, and d) the endogenous GTPase rate. Cytoskeleton provides kits for all these assays (e.g. BK124, BK100, BK100 and BK105).
- Measuring the active GTP-bound form of Rho: Traditionally, this assay has been performed using a pull-down method, wherein the Rho-GTP- binding domain (RBD) of a Rho effector is coupled to agarose beads, allowing affinity based detection of the active Rho in biological samples (11). This method suffers from several drawbacks such as being time consuming, requiring large amount of total cellular protein, being limited in the number of samples that can be handled simultaneously and yielding only semi-quantitative results. Cytoskeleton brought this concept to a new level of accuracy and sensitivity by introducing the GLISA technology which uses 96-well format and small amounts of cells/protein and provides highly accurate results. (more details are described here).
- Measuring total Small G-protein levels: Once assumed to be an insignificant bystander in cell signalling the total level of Small G-proteins has become a real factor in normal and diseased cell function (12, Burridge 2008). Using either a western blot approach (e.g. with ARH03), or a sandwich ELISA (e.g. BK150) it is possible to measure total Rho levels in a cell or tissue extract.
- Measuring GEF exchange activity: Using GTP fluorescent conjugates, which show differential fluorescence upon binding Small G-proteins, one can determine the rate of exchange of GTP, for more information see Cat. # BK100. This assay can be performed on purified recombinant proteins or by immunoprecipitating Rho protein from a cell extract (see the paper referenced in BK100 FAQ Question 1: BK100).
- Measuring endogenous GTPase activity: Although it is difficult to measure GTPase activity from cell extracts because of competing non-specific phosphatase activity, it is relatively straightforward to measure it from a purified recombinant protein with a phosphate assay such as Cat. # BK105 or BK054 offered by Cytoskeleton.
Guanine exchange factors
GEFs catalyze the exchange of GDP for GTP to generate the active state of small GTPases in response to extracellular signals. In order to facilitate the exchange, the GEFs must bind to the GDP bound GTPases, destabilize the GDP-GTPase complex, and then stabilize a nucleotide free reaction intermediate. Because of the high intracellular ratio of GTP to GDP, the released GDP is replaced with GTP, leading to release of GEFs from the complex and activation of the GTPase (1,2). Many GEF proteins have been identified as oncogenes and are involved in human disease such as cancer. Interestingly, the expression of GEF protein is tissue or cell type specific, providing a therapeutic potential for cancer treatment (2, 13).
Recently developed fluorescence analogs of guanine nucleotides have greatly improved the technical ability to define the real time exchange reaction of GEFs, including kinetic and thermodynamic properties, eliminating the need for traditional radioactive labeling method (14, 15). This fluorescence based assay takes advantage of the spectroscopic difference between bound and unbound fluorescent analogs to guanine nucleotides and thus is able to monitor the states of small GTPases (14, 15). One of most widely used fluorescent analogs is mant (N-methylanthraniloyl) fluorophores. Excitation of mant flurophore at 360nm (+/-10nm) gives a fluorescence emission at 440nm (+/-20nm). Once bound to GTPases, the fluorophore emission intensity increases dramatically approximately 2 fold. Therefore, the enhancement of fluorescent intensity in the presence of small GTPases and GEFs will reflect the respective GEF activities of known or unknown proteins.
Cytoskeleton Inc. has developed a Mant fluorophore based GEF assay suitable for both 96-well and 384-well format. This assay can be applied to multiple research purposes such as characterizing the GEFs and identifying GEF inhibitors in a high throughput screen format. This kit contains human Cdc42, Rac1 and RhoA proteins and the GEF domain of Dbs as a positive control GEF for Cdc42 (fig. 1) and RhoA. Dbs shows extremely low GEF activity for Rac1 (see Fig.2 and ref. 4). Interestingly, it was reported that human Dbs can activate Rac1 in a FRET based assay (16).
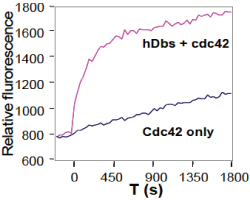
Legend: The small GTPases Cdc42 (Cat# CD01) and the human Dbs protein (Cat# GE01) were expressed as His-tagged proteins in E. coli and purified on a nickel affinity column. The reactions were conducted in a 96 well black flat bottom half area plate (Corning Cat# 3686) format (150 µl reaction volumes). Each reaction contains 1 µM GTPases, 50 µg/ml bovine serum albumin (BSA), 20 mM Tris pH 7.5, 50 mM NaCl, 10 mM MgCl2, and 0.8 µM mant-GTP with or without the presence of 0.5 µM human Dbs(DH/PH) protein. Reactions were measured in a Tecan Spectrofluor plus fluorimeter(lex=360nm, lem=460nm). Readings were taken at 20°C every 30 seconds for a total reaction time of 30 minutes.
GTPase Activating Proteins
The balance of the GTP to GDP bound state underlies the switch mechanism as they turn from an activated (GTP form) to inactive state (GDP form). The balance of GTP to GDP bound states is controlled by catalytic proteins that either increase the rate of exchange of GDP for GTP (GEFs), increase the GTPase activity (GAPs), or prevent the exchange of GDP (GDIs). Recently it has been shown that the GAP family of proteins is large (70 members) and potentially important for changing a cell from a normal to disease status (see review in ref. 17).
GAP activity can be reduced by deletion or mutation in cells; in this case the small GTPase targeted by the GAP has an extended time in the active GTP state, in essence creating a permanently active state. For example the ability of Rho GTPase to control axonal guidance is exemplified in the finding that mutations of a Rho-GAP can lead to mental retardation (18,19). In cancer, mutated Ras-GAPs (NF1 gene) have been shown to cause neurofibromatosis (20,21), and mutated Rheb-GAP has been shown to cause tuberous sclerosis complex (21).
Although some GAPs are known to have GAP activity, the majority of assumed GAP proteins have only been implicated by homology to contain GAP activity. Cytoskeleton Inc. is facilitating the exploration of this field by introducing the GAP Assay Kit (Cat. # BK105). Several small GTPase proteins (Ras, RhoA, CDC42 and Rac1) are included such that the researcher can screen the small G-proteins for GAP like activity which is usually small G-protein specific i.e. RhoGAP or Ras GAP. It is likely that new domain information will identify other small G-protein GAPs which have not been apparent. In addition the new assay kits are easily adapted for High Throughput Screen format which allows development of ligands for pharmaceutical studies. Please inquire for bulk reagent quotes if you are considering a screening application (tservice@cytoskeleton.com).
The reagents in this kit have been optimized to enable high activity from your GAP protein such that you can detect the enhanced GTPase of a small G-protein through a simple absorbance based detection method. The small G-protein is incubated in the presence of GAP protein (Rho GAP is the control protein included in this kit), GTP and the optimized buffer. The overall GTPase activity of small G-proteins is composed of two components which limit the activity; these are a) endogenous GTPase activity and b) GDP dissociation rate. The endogenous activity can be enhanced by the addition of a suitable GAP protein, whereas the dissociation rate can be enhanced by buffer optimization.
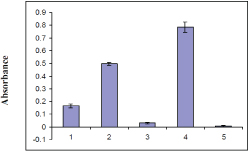
Figure 2. p50 RhoGAP activity measured by GTP hydrolysis by RhoA and Rac1 protein
References
- Shielge, J. M., et al. Trend Cell Biol., 2000, 10, 147-54.
- Whitehead, I. P., et al. Biochim. Biophys. Acta, 1997, 1332, F1-23.
- Jaffe, AB. & Hall, A. Rho GTPases: Biochemistry and Biology. Ann. Rev. Cell Dev. Biol. 21: 247-269 (2005)
- Ridley, AJ. & Hall, A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70: 389-399 (1992)
- Ridley, AJ. et al. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70: 401-410 (1992)
- Coso, OA., et al. The small GTP-binding proteins Rac and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81: 1137-1146 (1995)
- Small, JV., et al. The lamellipodium: where motility begins. Trends Cell Biol. 12: 112-120 (2002)
- Jaffe, A, & Hall, A, Rho GTPases in transformation and metastasis, Adv. Cancer Res. 84: 57-80 (2002)
- Chang L. et al. 2007. Rad GTPase Deficiency Leads to Cardiac Hypertrophy. Molecular Cardiology, 116: 2976-2983.
- Clark et al. 1993. Selective amplification of novel members of the ADF-ribosylation factor (Arf) family: cloning of new human and Drosophila ARL genes. PNAS, 90, 8952-6.
- Herrmann, C., Martin, G.A. and Wittinghofer, J. Biol. Chem. 270: 2901 (1995)
- Burridge Rho levels ref.
- Zheng, Y. Trend Biochem Sci.2002. 26, 724-32.
- Cheng, L., et al. Mol. Cell. Biol. 2002. 22, 6895-6905.
- Rossman, K. L., et al. Embo J. 2002. 21, 1315-1326.
- Whitehead, I. P., et al. Mol. Cell. Biol. 1999. 19, 7759-7770.
- Bernards A. and Settleman J. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14, (7), 377-385.
- Billuart P. et al. 1998. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature, 392, 923-926.
- Faucherre A. et al. 2003. Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum. Mol. Genet. 12, 2449-2456.
- Cichowski K. and Jacks T. 2001. NF1 tumor suppressor gene function: narrowing the GAP. Cell, 104, 593-604.
- Li Y. et al. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29, 32-38.
