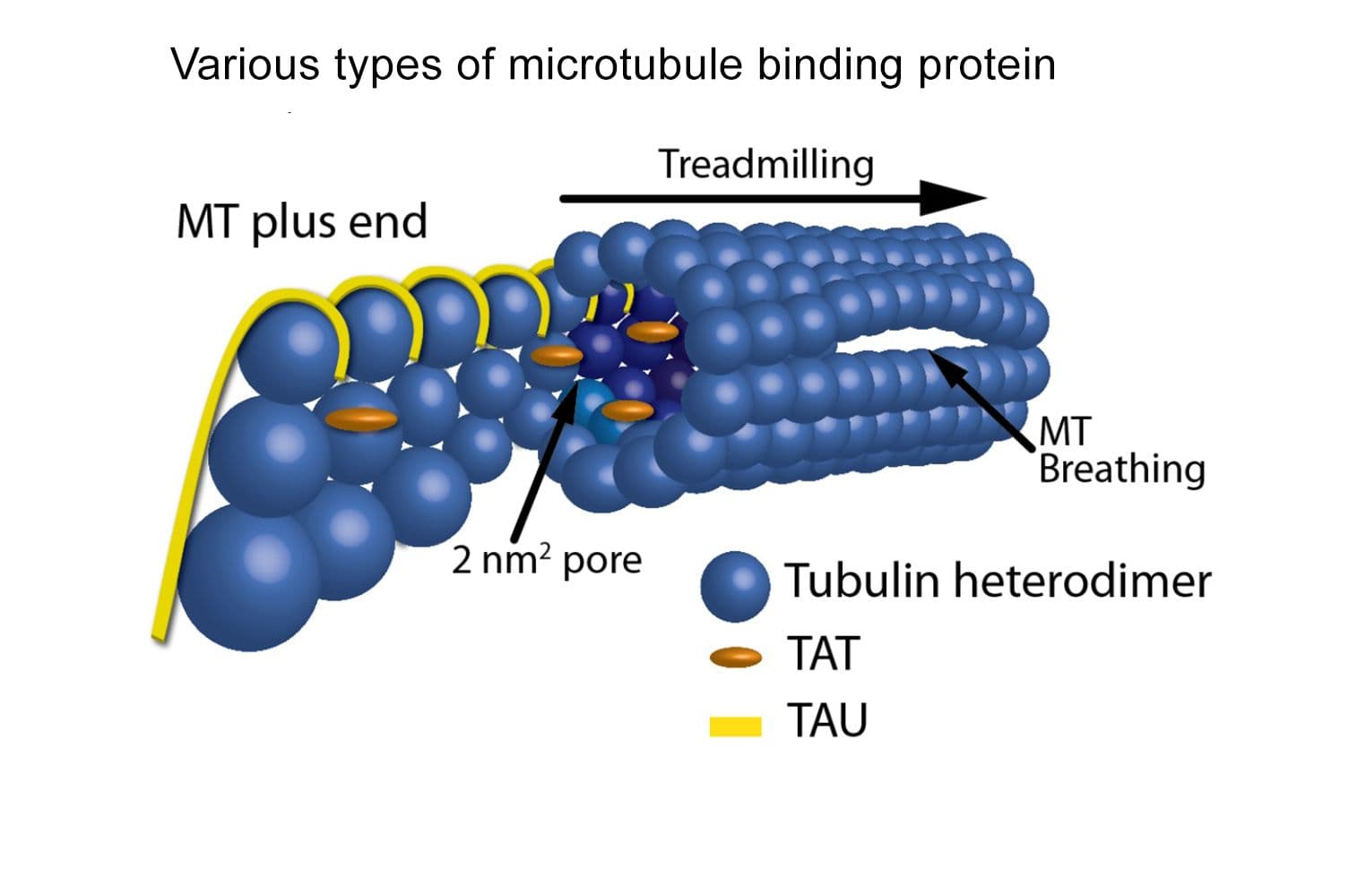
+3
Kit content (50 - 100 assays)
Equipment & materials required
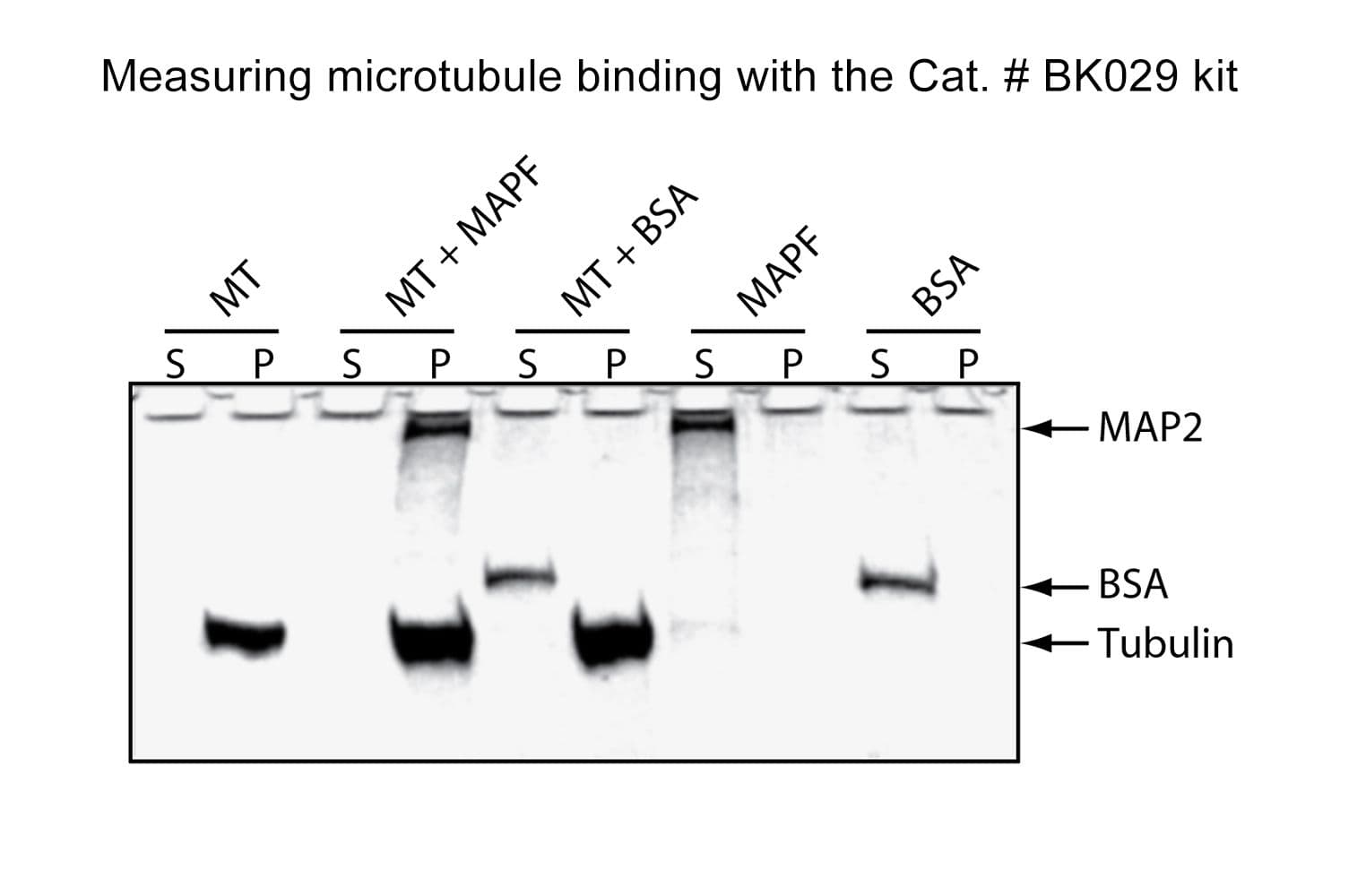
This assay takes advantage of the fact that microtubules (MTs) pellet when centrifuged at 100,000 × g. Any protein bound to MTs will co-pellet during centrifugation. When using purified proteins, comparing the supernatant and pellet fractions via SDS-PAGE is sufficient to determine MT association. When working with cell lysates or in vitro translation products, detection can be performed using western blotting.
Key characteristics
Cat. #BK029