Monoubiquitination: A dynamic tag for protein regulation
Ubiquitination is a post-translational modification (PTM) that involves the covalent attachment of an 8 kDa ubiquitin (Ub) peptide to one or more lysines of a target protein. Modification of a target protein may occur as a single Ub on a single lysine (monoubiquitination), a single Ub on multiple lysines (multiubiquitination) or as ubiquitinated chains in which lysines on the initial protein-conjugated ubiquitin are extended through sequential rounds of ubiquitination (polyubiquitination). The fact that ubiquitin contains seven lysine residues and polyubiquitination has been demonstrated to occur through all of these gives this PTM the potential for highly diverse modifications of target proteins1. It is also known that ubiquitination, through the action of deubiquitinating proteins (DUBs), is a rapidly reversible process2, thus giving ubiquitination the potential to be a hugely versatile signaling moiety, far exceeding the complexity of more well-studied PTMs such as phosphorylation. While the critical role of ubiquitination in the proteasomal degradation of target proteins is well known3, the non-degradative roles of ubiquitination are only beginning to be described. This newsletter focuses on several general, non-degradative processes regulated by monoubiquitination (Fig. 1).
Intramolecular Autoinhibition
PTMs such as phosphorylation are major regulators of protein autoinhibition4. Evidence is emerging to suggest that protein monoubiquitination may serve a similar function. Several reports have shown that proteins containing ubiquitin binding domains (UBDs) are often monoubiquitinated (mUb), thereby suggesting a simple autoinhibitory mechanism via an intramolecular UBD-mUb interaction5,6. Endocytic adaptor proteins such as Sts1, Sts2, and Eps15 provide the best examples of this type of regulation6,7. Receptors, such as the epidermal growth factor receptor (EGFR), become ubiquitinated post-activation, which triggers internalization and degradation through the endosomal-lysosomal pathway, thereby attenuating the transduction signal. The endocytic adaptor proteins control the internalization and degradation of ubiquitinated cell surface receptors by binding the receptor’s Ub moiety in trans via the adaptor UBD domain and linking the receptor to endosomal trafficking proteins. In cases where the adaptor proteins are autoinhibited, and therefore blocked from recognizing their ubiquitinated receptor target, the transduction signal will continue to be propagated. In these cases, autoinhibition acts as a modulator of receptor-mediated signal transduction7 (Fig. 1).
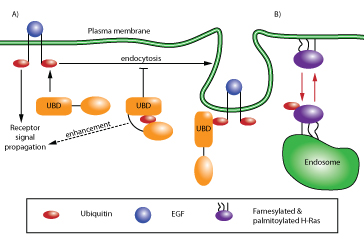
Figure 1: Schematic of ubiquitin-mediated signaling and trafficking. (A) Pathway denoted by black arrows illustrates ubiquitin-driven autoinhibition. Autoinhibition of the UBD-containing protein regulates receptor endocytosis and signal transduction (e.g., EGFR recycling7). (B) Pathway denoted by red arrows illustrates ubiquitin-regulated cell localization of a protein. This type of regulation occurs with H- and N-Ras, where ubiquitination localizes Ras from the plasma membrane to the endosomal membrane13.
There are twenty known families of UBDs and greater than 150 proteins that contain one or more UBD domains8. The UBD proteins are involved in a wide range of cellular processes, including endocytosis, DNA repair, transcriptional regulation, and signal transduction6. Current evidence suggests that ubiquitin-mediated autoinhibition may play a major role in regulating these processes.
Intracellular Localization and Trafficking
Proper cellular localization of a protein is a primary means of regulating its activity and many strategies have evolved to ensure the fidelity of spatial regulation9, including well-documented cases of PTM events such as farnesylation, palmitoylation, and phosphorylation10-12. Monoubiquitination can act in conjunction with other PTMs to localize proteins such as small GTPases, cytoskeletal proteins, and scaffolding proteins to specific cellular compartments (Fig. 1). For example, the isoforms H-Ras, N-Ras, and K-Ras have all been shown to be substrates for monoubiquitination (and non-degradative diubiquitination in the case of H- & N-Ras)13,14. H-Ras ubiquitination appears to act in conjunction with farnesylation and palmitoylation to promote relocation of H-Ras from the plasma membrane to endosomal sites with a concomitant reduction in MAPK signaling13. Interestingly, monoubiquitination of K-Ras, the Ras gene most commonly associated with cancer, appears to enhance GTP loading by modulating the affinity of K-Ras for downstream effectors15. Similarly, signaling in response to VEGF also involves monoubiquitination. The monoubiquitinated form of the actin binding protein filamin B has been shown to regulate nuclear/cytoplasmic trafficking and cytoplasmic localization of HDAC7 in response to VEGF16. Transient monoubiquitination of filamin B occurs in response to VEGF stimulation and binds to the nuclear localization signal (NLS) of HDAC7, thereby transiently preventing its re-entry into the nucleus and mitigating the transcriptional repressor activity of HDAC7 on the genes required for VEGF-mediated responses16. Another structurally-related protein targeted for monoubiquitination is the Toll-like and interleukin-1 receptor complex scaffold protein TRAF4. This protein undergoes monoubiquitination as a prerequisite for localization at tight junctions where it is required for Rac1 activation and migration of normal breast epithelial and cancer cells17.
Regulation of Protein Complex Formation
Monoubiquitination has been reported to regulate protein complex formation in a number of systems18,19. For example, the process of transcriptional elongation is known to require nucleosomal rearrangement to remove the physical block from the transcribing RNA polymerase. The FACT (facilitates chromatin transcription) complex is a major player in this process and operates, in part, by displacing a histone H2A/H2B dimer from the nucleosome. An in vitro reconstitution system demonstrated that monoubiquitination of H2B is required for FACT-mediated displacement of the H2A/H2B dimer18.
In conclusion, monoubiquitination, and non-degradative ubiquitination signaling in general, is rapidly becoming established as a versatile regulatory motif.
Related Research Tools
| Signal-Seeker™ Kits | Cat. # | Amount | ||||||
Signal-Seeker™ Ubiquitination Detection Kit 30 assay | BK161 | 30 assays | ||||||
Signal-Seeker™ Ubiquitination Detection Kit 10 assay | 10 assays | |||||||
| Signal-Seeker™ Ubiquitination detection reagents | Cat. # | Amount | ||||||
Ubiquitination Affinity Beads | UBA01-Beads | 40 reactions | ||||||
Ubiquitination Control Beads | CUB02-Beads | 10 reactions | ||||||
Ubiquitin Antibody Mouse Monoclonal | AUB01 | 2 x 100 µl | ||||||
BlastR™ Rapid Lysate Prep Kit | BLR01 | 50 purifications | ||||||
References
- Sadowski, M. et al. 2012. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 64:136-142.
- Burrows, J. et al. 2012. Regulation of cellular responses by deubiquitinating enzymes: an update. Frontiers Biosci. 17:1184-1200.
- Chitra, S. et al. 2012. The ubiquitin proteasome system and efficacy of proteasome inhibitors in diseases. Int. J. Rheum. Dis. 15:249-260.
- Betschinger, J. et al. 2005. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein lethal (2) giant larvae. Cell. 15:276-282.
- Hicke, L. et al. 2005. Ubiquitin binding domains. Nature Rev. Mol. Cell Biol. 6: 610-621.
- Holler, D. et al. 2006. Regulation of ubiquitin binding proteins by monoubiquitination. Nature Cell Biol. 8:163-169.
- Fallon, L. et al. 2006. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 8:834-842.
- Dikic, I. et al. 2009. Ubiquitin-binding domains: from structures to functions. Nature Rev. Mol. Cell Bio. 10:659-671.
- Nabi, I.R. (Ed). 2011. Cellular Domains. Wiley-Blackwell Press: Hoboken, NJ.
- Basso, A. et al. 2005. Thematic review: lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 47:15-31.
- Salaun, C. et al. 2010. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191:1229-1238.
- York, B. et al. 2006. Tuberin nuclear localization can be regulated by phosphorylation of its carboxyl terminus. Mol. Cancer Res. 4:885-897.
- Jura N. et al. 2006. Differential modification of Ras proteins by ubiquitination. Mol. Cell. 21:679-687.
- Ahearn I., et al. 2012. Regulating the regulator: post-translational modification of Ras. Nature Rev. 13:39-51.
- Sasaki, A. et al. 2012. Ubiquitination of Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 4:ra13.
- Su, Y. et al. 2013. Monoubiquitination of filamin B regulates vascular endothelial growth factor-mediated trafficking of histone deacetylase 7. Mol. Cell Biol. 33:1546-1560.
- Wang X. et al. 2013. Ubiquitination of tumor necrosis factor receptor-associated factor 4 (TRAF4) by SMAD ubiquitination regulatory factor 1 (Smurf1) regulates motility of breast epithelial and cancer cells. J. Biol. Chem. 288:21784-21792.
- Pavri, R. et al. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 125:703-717.
- Bienko, M. et al. 2010. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Cell. 37:396-407.

