July Newsletter: Post-translational Regulation of Mdm2, an E3 Ubiquitin Ligase for Tumor Suppressor p53
The ubiquitin E3 ligase Mdm2 (murine double minute 2; human homolog, Hdm2) is well known for its oncogenic activities and as the master regulator of the powerful tumor suppressor p531,2. Moreover, Mdm2 may function as an oncogenic protein independent of p533. Mdm2 is able to inhibit p53-mediated gene expression through two pathways: inhibition of transcriptional activity by direct binding and ubiquitin-mediated degradation via its E3 ligase activity4; however, the effectiveness of p53 inhibition by direct Mdm2 binding has been questioned5.
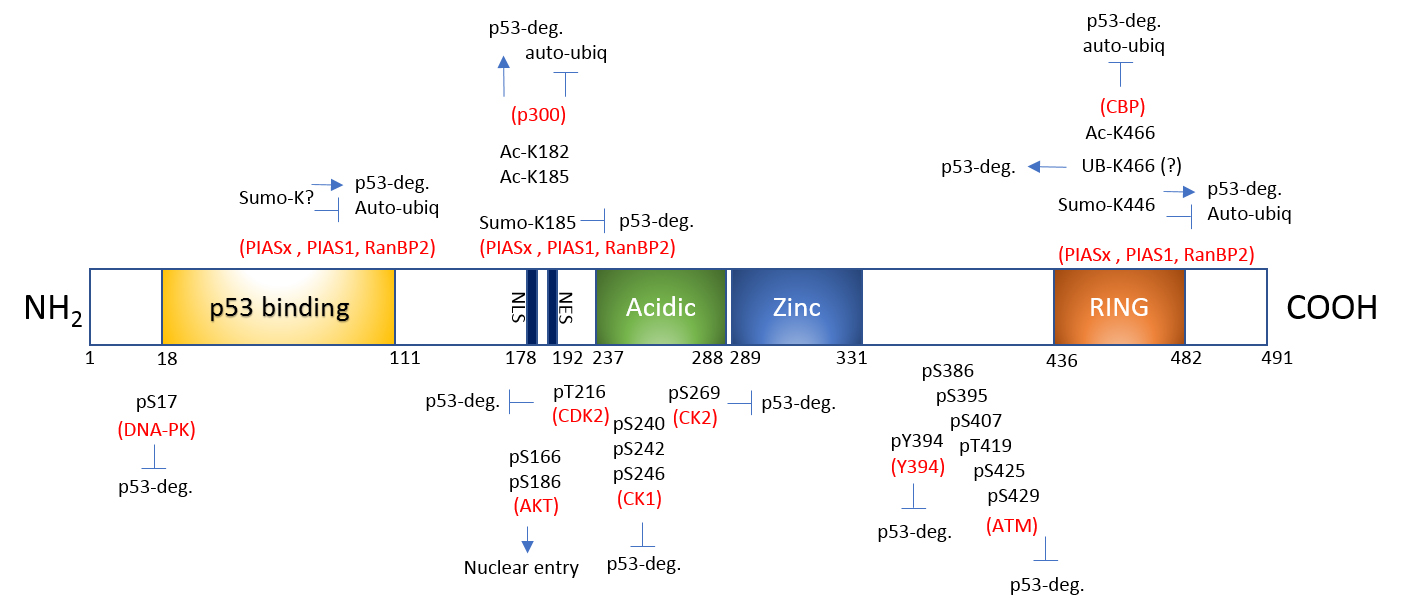
Under normal, non-stress conditions, Mdm2 maintains p53 expression and activity at a minimal level to tightly regulate its apoptotic/cell death transcriptional activities. Under conditions of cellular stress, Mdm2-mediated ubiquitination of p53 ceases, allowing p53 to activate transcription of apoptotic genes and those involved in inhibiting cell growth. Much research concludes this is due to Mdm2's auto-inhibition by self-ubiquitination1-3. However, this story appears to be more complex than originally thought5 and involves multiple post-translational modifications (PTMs). Here, we discuss Mdm2's regulation by ubiquitination, SUMOylation, phosphorylation, and acetylation6-8 (Fig. 1).
Ubiquitination and SUMOylation of Mdm2
Mdm2 is ubiquitinated, at least partially by Mdm2 itself (auto-ubiquitination, common to most E3 ligases), which leads to its destabilization and degradation1-3 (Fig. 1). However, Mdm2's E3 ubiquitin ligase activity is not necessary for Mdm2 degradation, suggesting that auto-ubiquitination is not the only means by which Mdm2 is degraded5. There are other candidate proteins with E3 ligase activity such as p300-CBP-associated factor (PCAF) which ubiquitinates Mdm2 and regulates its stability9. The role of ubiquitination in regulating Mdm2 activity is further complicated with the report that poly-ubiquitination of Mdm2 activates its E3 ligase activity as this modification significantly enhanced the capacity to poly-ubiquitinate p5310. Interestingly, poly-, but not mono-, ubiquitination upregulated Mdm2's E3 ligase activity10. These findings challenge the assumption that auto-poly-ubiquitination only signals degradation10.
SUMOylation confers proteins with differential stability, localization, inter-protein interaction, and DNA binding/transactivation11. Mdm2 is SUMO1-modified by SUMO E3 ligases Ubc9, PIAS1, and RanBP212,13 (Fig. 1). Upon nuclear entry, Mdm2 is SUMO1-modified by RanBP2 and further SUMOylated by PIAS, which favors p53 stability and abundance in the nucleus13. This modification can be reversed by stress induction as UV irradiation decreases Mdm2 SUMOylation12. SUMOylated Mdm2 minimizes auto-ubiquitination and maximizes its E3 ligase activity toward p5312. This is consistent with the idea that SUMO and ubiquitin modifications may be mutually exclusive and antagonistic.
Multi-site Phosphorylation of Mdm2
Serine and threonine residues constitute almost 20% of Mdm2's amino acids and many of these are phosphorylated by multiple kinases (Fig. 1). They are heavily clustered in two distinctive sites, the N-terminal and Central domains14. The phosphorylation of Mdm2 fine-tunes mainly the interaction with p53. Ataxia-telangiectasia mutated (ATM) kinase-mediated phosphorylation of Mdm2's S395 residue destabilizes Mdm2, thus reducing p53 degradation. Conversely, PI3-K/Akt-mediated phosphorylation of S166 and S186 residues stabilizes Mdm2 and destabilize p533,6,15 (Fig. 1). Upon genotoxic stress, DNA-dependent protein kinase (DNA-PK) phosphorylates the S17 residue of Mdm2, which dissociates p53 from Mdm216. As might be expected, protein phosphatase 1D (PPM1D; a.k.a. wild-type p53-induced phosphatase 1)-mediated dephosphorylation of phosphorylated S395 stabilizes Mdm2 and facilitates p53 degradation17 (Fig. 1).
Acetylation of Mdm2
Mdm2 is also acetylated by CREB-binding protein (CBP) and p300 in vitro and CBP in vivo, and the acetylation primarily occurs in the C-terminal RING finger domain8,18. These data suggest that acetyltransferases may positively modulate cellular p53 activity indirectly by inactivating Mdm2. In vitro acetylation by p300 also occurs within a central domain (124-246 amino acids) of Mdm218. More recently, acetylation on two lysine residues (K182 and K185) in the nuclear localization sequence has been identified7 (Fig. 1). Under normal, non-stress conditions, Mdm2 is acetylated by p300 on K182 and K185, facilitating binding of the deubiquitinase HAUSP (herpes virus-associated ubiquitin-specific protease) to Mdm2. These modifications (acetylation, presumed de-ubiquitination) stabilize Mdm2 and protect it from auto-ubiquitination, which correlates with increased inhibition of p53 activity. Genotoxic stress activates the deacetylase SIRT1 and SIRT1-mediated deacetylation of these sites promotes Mdm2 auto-ubiquitination, thereby increasing p53 stability and subsequent activity7,19. Therefore, p300-mediated acetylation and SIRT1-mediated deacetylation on K182 and K185 regulate the functional stability of Mdm2 (with help from HAUSP) and serve as molecular switches for control over p53 activity7 (Fig. 1).
Summary
PTMs are well known regulators of a protein's function, localization, and stability. In the case of Mdm2, the ubiquitination, SUMOylation, phosphorylation, and acetylation modifications regulate Mdm2's function and stability both independently and via cross-talk with one another. In so doing, precise and quick Mdm2-mediated control over p53-stimulated cell death and survival is possible under various types and intensities of cellular stress. Cytoskeleton, Inc. offers many different Signal Seeker™ research tools for dissecting and analyzing the role that PTMs play in the regulation of virtually any protein, including clinically relevant proteins in neurobiology, cardiology, rheumatology, and oncology.
Related Products & Resources
Signal Seeker™ Kits
Signal-Seeker™ Acetyl-Lysine Enrichment Kit (Cat. # BK163)
Signal-Seeker™ Phosphotyrosine Enrichment Kit (Cat. # BK160)
Signal-Seeker™ SUMO 2/3 Enrichment Kit (Cat. # BK162)
Signal-Seeker™ Ubiquitin Enrichment Kit (Cat. # BK161)
PTM Antibodies, Beads, Etc.
Acetyl-Lysine antibody Mouse Monoclonal (7B5A1) (Cat. # AAC02)
Acetyl-Lysine antibody Mouse Monoclonal (19C4B2.1) (Cat. # AAC03)
Acetyl-Lysine-HRP antibody Mouse Monoclonal (19C4B2.1) (Cat. # AAC03-HRP)
Acetyl-Lysine Affinity Beads (Cat. # AAC04-beads)
Phosphotyrosine Affinity Beads (Cat. # APY03-beads)
SUMO 2/3 Affinity Beads (Cat. # ASM24-beads)
Ubiquitin Affinity Beads (Cat. # UBA01-beads)
Control for Ippt IgG Beads (Cat. # CIG01-beads)
Control for Ubiquitin Affinity Beads (Cat. # CUB02-beads)
References
- Karni-Schmidt O. et al. 2016. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617-644.
- Wade M. et al. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer. 13, 83-96.
- Iwakuma T. and Lozano G. 2003. MDM2, an introduction. Mol. Cancer Res. 1, 993-1000.
- Momand J. et al. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 69, 1237-1245.
- Itahana K. et al. 2007. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 12, 355-366.
- Meek D.W. and Knippschild U. 2003. Posttranslational modification of MDM2. Mol. Cancer Res. 1, 1017-1026.
- Nihira N.T. et al. 2017. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci. Signal. DOI: 10.1126/scisignal.aai8026.
- Wang X. et al. 2004. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett. 561, 195-201.
- Linares L. et al. 2007. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol. 9, 331-338.
- Ranaweera R.S. and Yang X. 2013. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J. Biol. Chem. 288, 18939-18946.
- Geiss-Friedlander R. and Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947-956.
- Buschmann T. et al. 2001. The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J. Biol. Chem. 276, 40389-40395.
- Miyauchi Y. et al. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277, 50131-50136.
- Hay T.J. and Meek D.W. 2000. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 478, 183-186.
- Mayo L.D. and Donner D.B. 2001. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 11598-11603.
- Mayo L.D. et al. 1997. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 57, 5013-5016.
- Lu X. et al. 2008. The Wip1 phosphatase and Mdm2: cracking the "Wip" on p53 stability. Cell Cycle. 7, 164-168.
- Kawai H. et al. 2001. Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 276, 45928-45932.
- Michael D. and Oren M. 2003. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13, 49-58.