June Newsletter: Posttranslational Regulation of Key Transcription Factors in Pluripotent Stem Cells
Pluripotent stem cells (PSCs), characterized by their unlimited self-renewal and differentiation potential, have garnered special attention as therapeutics because of their capacity to differentiate into any cells of the respective adult organism1. There are two general types of PSCs: 1. embryonic stem cells (ESCs) and 2. induced pluripotent stem cells (iPSCs). By definition, PSCs exist in a pluripotent state until differentiation into specialized cells. To maintain stem cells as pluripotent, select transcription factors activate pluripotency-promoting genes and concomitantly suppress differentiation-promoting genes. In turn, the expression level and transactivation ability of these transcription factors are regulated by post-translational modifications (PTMs)2,3. Three key pluripotent transcription factors are Oct4, Sox2, and Nanog3. Their regulation of transcription is complex with each transcription factor able to function independently of the other while also capable of auto-inhibition (e.g., Oct4)4

or forming a heterodimer whereby one factor is regulated by the complex (e.g., Nanog activity is strictly regulated by the Oct4/Sox2 heterodimer)5,6. Understanding the precise regulation of the stability (expression level) and transcriptional activity (DNA-binding affinity) of Oct4, Sox2, and Nanog by PTMs is essential in the study of stem cell homeostasis7. In this newsletter, we discuss how PTMs such as ubiquitination, SUMOylation, phosphorylation, methylation, and acetylation regulate the activities of the Oct4, Sox2, and Nanog pluripotency transcription factors (Fig. 1).
PTM regulation of Oct4
The expression level and transcriptional activity of Oct4 are regulated by ubiquitination, SUMOylation, and phosphoylation. Ubiquitination-proteasome degradation is the main control system for maintaining and monitoring the quality and levels of protein expression in cells. When Oct4 is ubiquitinated by the wwp2 E3 ligase on K63, the level of Oct4 is decreased and ESCs are induced to differentiate into a given cell type7,8. In addition to its expression level, the PTM status of Oct4 governs its transcriptional activity. SUMOylation of Oct4 on K118, near the Pit1-Oct-Unc86 (POU) DNA-binding domain of Oct4, significantly increases its stability as well as its DNA-binding affinity (transactivation potential)9 (Fig. 1). Oct4 is able to form heterodimers with other transcription factors or form homodimers, which diversifies its potential transcriptional activities10. Phosphorylation of Ser229 by protein kinase A blocks Oct4 homodimer conformation and hinders sequence-specific DNA binding11. Also, phosphorylation of T234 and S235 residues by an unknown kinase(s) within the POU DNA-binding domain negatively regulates Oct4's activity12 (Fig. 1). Phosphorylation on the Y327 residue by Abl kinase might influence the DNA binding and transcriptional activity of Oct4, but this has yet to be determined12.
PTM regulation of Sox2
The expression level of Sox2 in ESCs is precisely regulated by a competition between methylation and phosphorylation13. Methyltransferase Set7 mono-methylates Sox2 at K119, which inhibits transcriptional activity and enhances Sox2 ubiquitination (Fig. 1). In contrast to the inhibition of transcriptional activity, the phosphorylation on T118 by AKT1 stabilizes Sox2 and maintains the expression level by inhibiting the ubiquitination-proteasome pathway. Phosphorylation of three Sox2 serine residues (S249, S250, and S251) facilitates its SUMO modification (K245)14 (Fig. 1). Unlike in the case of Oct4, the SUMOylation of Sox2 impairs DNA binding and negatively regulates its transcriptional activity15. It is a reasonable speculation that the phosphorylation-dependent SUMOylation is a nuclear import mechanism, considering that SUMOylation increases the stability and the trafficking of proteins into the nucleus in a stress situation16. Acetylation of Sox2 at K75 by histone acetyltransferase p300/CBP enhances nuclear export and the degradation of Sox2 through a ubiquitin-mediated degradation pathway17 (Fig.1 ).
PTM regulation of Nanog
Nanog undergoes phosphorylation on multiple serine residues by ERK1, which reduces its activity levels and in turn enhances ESC differentiation18. The phosphorylation by ERK1 makes Nanog susceptible to ubiquitination by the E3 ubiquitin ligase FBXW8, which decreases its stability. Conversely, ubiquitin-specific peptidase21 (USP21) interacts with Nanog in ESCs and de-ubiquitinates a K48 chain-specific ubiquitin linkage in order to regulate Nanog's stability and maintain the pluripotential state of ESCs19 (Fig. 1).
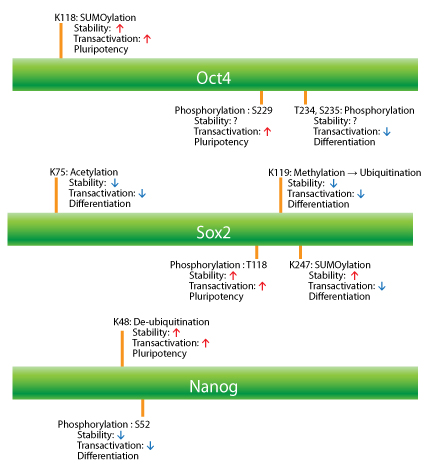
SUMOylation of Nanog has not been reported yet, but Nanog's expression is regulated by the SUMOylation of Oct4 and Sox2 in an opposite manner; SUMOylation of Oct4 up-regulates Nanog expression, while SUMOylation of Sox2 down-regulates Nanog expression20.
Summary
The expression levels and transactivation of pluripotency transcription factors, which determine the stem cell fate of pluripotency versus differentiation, are strictly regulated by PTMs. As a fine-tuning mechanism, various PTMs function antagonistically or co-operatively in the pluripotency regulatory circuits governing the activities of Oct4, Sox2, and Nanog. Thus, the precise and accurate identification of PTMs on individual stem cell protein markers is of paramount importance in understanding how these transcription factors interact with each other and regulate stem cell lineage. To aid scientists in identifying the PTMs of stem cell proteins and characterizing novel PTM-mediated regulation of pluripotency transcription factors, Cytoskeleton, Inc. offers Signal SeekerTM kits that reveal the PTM status of pluripotency factors from a simple stem cell lysate. PTMs such as tyrosine phosphorylation, ubiquitination, SUMOylation, and acetylation can be identified and quantified.
Related Products & Resources
Signal Seeker™ Kits
Signal-Seeker™ Phosphotyrosine Enrichment Kit: 30 Assays (Cat. # BK160)
Signal-Seeker™ Phosphotyrosine Enrichment Kit: 10 Assays (Cat. # BK160-S)
Signal-Seeker™ Ubiquitin Enrichment Kit: 30 Assays (Cat. # BK161)
Signal-Seeker™ Ubiquitin Enrichment Kit: 10 Assays (Cat. # BK161-S)
Signal-Seeker™ SUMO 2/3 Enrichment Kit: 30 Assays (Cat. # BK162)
Signal-Seeker™ SUMO 2/3 Enrichment Kit: 10 Assays (Cat. # BK162-S)
PTM Antibodies, Beads, Etc.
Phosphotyrosine Affinity Beads (Cat. # APY03-beads)
SUMO 2/3 Affinity Beads (Cat. # ASM24-beads)
Ubiquitin Affinity Beads (Cat. # UBA01-beads)
Control for Ippt IgG Beads (Cat. # CIG01-beads)
Control for Ubiquitin Affinity Beads (Cat. # CUB02)
Spirochrome™ Live Cell Imaging Probes
SiR-Actin Kit, Cat. # CY-SC001
SiR700-Actin Kit, Cat. # CY-SC013
SiR-Tubulin Kit, Cat. # CY-SC002
SiR700-Tubulin Kit, Cat. # CY-SC014
Cytoskeleton Kit (SiR-Actin and SiR-Tubulin), Cat. # CY-SC002
SiR-DNA Kit, Cat. # CY-SC007
SiR700-DNA Kit, Cat. # CY-SC015
SiR-Lysosome Kit, Cat. # CY-SC012
SiR700-Lysosome Kit, Cat. # CY-SC016
References
- Trounson A. et al. 2016. Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194-200.
- Young R.A. 2011. Control of the embryonic stem cell state. Cell. 144, 940-954.
- De Los Angeles A. et al. 2015. Hallmarks of pluripotency. Nature. 525, 469-478.
- Pan G. et al. 2006. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 20, 1730-1732.
- Boyer L.A. et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122, 947-956.
- Rodda D.J. et al. 2005. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731-24737.
- Cai N. et al. 2012. Post-translational modulation of pluripotency. J. Mol. Cell Biol. 4, 262-265.
- Xu H.M. et al. 2004. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279, 23495-23503.
- Wei F. et al. 2007. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551-21560.
- Reményi A. et al. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048-2059.
- Saxe J.P. et al. 2009. Post-translational regulation of Oct4 transcriptional activity. PLoS One. 4, e4467.
- Brumbaugh J. et al. 2012. Phosphorylation regulates human OCT4. Proc. Natl. Acad. Sci. U.S.A. 109, 7162-7168.
- Fang L. et al. 2014. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell. 255, 537-551.
- Van Hoof D. et al. 2009. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 5, 214-226.
- Tsuruzoe S. et al. 2006. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem. Biophys. Res. Commun. 351, 920-926.
- Geiss-Friedlander R. et al. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947-956.
- Baltus G.A. et al. 2009. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells. 27, 2175-2184.
- Kim S.H. et al. 2014. ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Res. 13, 1-11.
- Liu X. et al. 2016. USP21 deubiquitylates Nanog to regulate protein stability and stem cell pluripotency. Signal Transduct. Target. Ther. 1, e16024.
- Wu Y. et al. 2012. SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2. PLoS One. 7, e39606.
