Post-translational Modifications - Essential for Protein Regulation
To maintain homeostasis, cells need to respond to changes in the intracellular and extracellular milieu. Some of the changes have to be acted upon quickly to avoid detrimental effects that can lead to cell damage or even death. One way that cells act is through protein post-translational modifications (PTMs) which enable cells to respond quickly to various changes by influencing numerous properties of proteins including enzymatic activity, protein interactions, and subcellular location. Advanced developments in mass-spectrometry (MS) methods allow the identification of PTMs on a large scale. Today, more than 200 PTMs are known1,2, ranging from small chemical modifications (e.g., phosphorylation and acetylation) to the addition of complete proteins (e.g., ubiquitination and SUMOylation). The most extensively studied PTM is phosphorylation, in which protein kinases attach a phosphate group to serine, threonine, and/or tyrosine residues. Phosphorylated proteins appear to play a crucial role in many diverse and important cellular processes. Acetylation of histone proteins was discovered about 50 years ago3 and has long been associated with regulation of chromatin structure4. Several proteomics studies identified almost 5000 acetylation sites across almost 2000 proteins5,6. Many of them are localized in cytoplasm, mitochondria, and the plasma membrane, indicating their involvement in diverse cellular processes such as signal transduction, cytoskeletal dynamics, and membrane stability5,6 . In addition to acetylation, the lysine residue has been found to be modified by ubiquitination and SUMOylation, where the former plays a prominent role in protein degradation through proteasomal pathway7 and the latter is important for regulating cellular processes, including cell cycle, apoptosis, DNA repair, and signal transduction pathways8.

Fig. 1. Enrichment of tyrosine-phosphorylated proteins was performed by incubating APY03-beads with lysates from NIH3T3 cells either treated or untreated with hydrogen peroxide-activated sodium pervanadate. Western blot analyses using PTMtrue™ antibodies detect co-modified proteins (1B-1D) from the pY enrichment pool.
PTM crosstalk – Adding layers of complexity to the proteomic network
PTM crosstalk has gained increasing interest in recent proteomics studies. Two independent groups employed bioinformatics and database analyses of protein sequences and have identified “hotspots” where PTMs of different types are located within a close proximity, about 15 amino acids apart9,10. Some of these “hotspots” have been reported in the literature to crosstalk1,2. With the advancement in enrichment methods and MS technologies, several groups have conducted global proteomic analyses of PTMs in a cellular context11-13 and obtained evidence of crosstalk between phosphorylation and ubiquitination in protein degradation11 and SUMOylation-regulated protein phosphorylation (e.g., casein kinase II)12. PTM crosstalk can be either positive or negative in nature. In positive crosstalk, one PTM can serve as a signal for the addition or removal of a second PTM, or for recognition by a binding protein that carries out a second modification (e.g., phosphorylation-dependent SUMOylation)14. For negative crosstalk, it can be illustrated through a direct competition for modification of a single residue in a protein15,16, or indirectly by masking the recognition site for a second PTM15,16. Also, there are many examples showing that multiple modifications are present on a single protein1. These combinatorial actions of PTMs further expand the landscape of proteomes in eukaryotes, providing a fine-tuning mechanism in regulating protein function, localization, and interaction with other molecules.
Cytoskeleton, Inc. offers PTMtrue™ antibodies to assist researchers in studying PTMs of their target protein(s). We have recently developed PTM enrichment beads by cross-linking anti-phosphotyrosine (pY) antibody (Cat. # APY03) and anti-SUMO-2/3 antibody (Cat. # ASM24) to protein G beads. Figures 1 and 2 demonstrate the use of our PTM enrichment beads to probe for proteins co-modified with different PTMs. To enrich for tyrosine-phosphorylated proteins, lysates from NIH3T3 cells either treated or untreated with sodium pervanadate were incubated with APY03-beads. Enrichment of SUMO-2/3 targets was performed by incubating ASM24-beads with lysates from HeLa cells with or without heat shock treatment. Enriched proteins were eluted from beads and resolved on SDS-PAGE gels. Proteins were then transferred to PVDF membranes and PTMtrue™ antibodies, anti-pY (Cat. # APY03), anti- SUMO-2/3 (Cat. # ASM23), anti-Ubiquitin (Ub; Cat. # AUB01), and anti-acetyl lysine (Ac-K; Cat. # AAC01) were used to detect proteins with specific PTMs in the enrichment pools. As shown in Figures 1A and 2A, both of our enrichment beads exhibit superior pull-down abilities toward their target PTM proteins. We can also detect co-modified proteins from the pY and SUMO-2/3 enrichment pools using our PTMtrue™ antibodies (Figs. 1B-D; 2B-D).
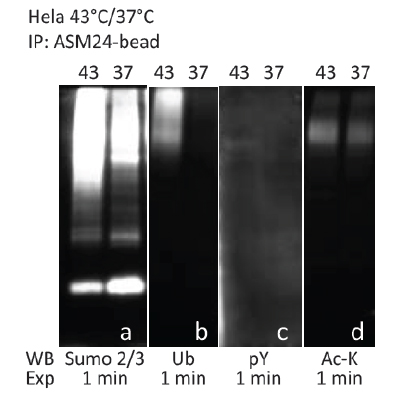
Fig. 2. Enrichment of SUMO-2/3 targets was performed by incubating ASM24-beads with lysates from HeLa cells after +/- heat shock treatment (43°C). Western blot analyses using PTMtrue™ antibodies detect co-modified proteins (2B-2D) from the SUMO-2/3 enrichment pool.
Conclusions
There are 2 bottlenecks in studying PTM crosstalk. First, PTMs often exist at sub-stoichiometric levels and require highly specific enrichment methods. Second, instead of intact proteins, peptides from trypsin-digested proteins are commonly used in proteomics study, which often severs the connection between multiple modifications present on a single protein isoform. Cytoskeleton, Inc. is excited to present a line of highly specific PTM antibodies to detect and enrich PTM proteins at the endogenous level. Please contact our Technical Service team (tservice@cytoskeleton.com) to see how we can support your research into PTMs.
PTMtrue™ Antibodies and Reagents
Anti-Acetyl Lysine Mouse Monoclonal Antibody (Cat. # AAC01)
Anti-Phosphotyrosine Mouse Monoclonal Antibody (Cat. # APY03)
Anti-Phosphotyrosine-HRP Mouse Monoclonal Antibody (Cat. # APY03-HRP)
Anti-SUMO 2/3 Mouse Monoclonal Antibody (Cat. # ASM23)
Anti-SUMO 2/3 Mouse Monoclonal Antibody (Cat. # ASM24)
New: SUMO 2/3 Affinity Beads (Cat. # ASM24-Beads)
Anti-Ubiquitin Mouse Monoclonal Antibody (Cat. # AUB01)
Live Cell Imaging Probes
Cytoskeleton Kit (Includes SiR-Actin, SiR-Tubulin, and Verapamil) (Cat. # CY-SC006)
SiR-Actin Kit (Cat. # CY-SC001)
SiR-Tubulin Kit (Cat. # CY-SC002)
References
- Minguez P. et al. 2012. Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. DOI: 10.1038/msb.2012.31.
- Beltrao P. et al. 2013. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. DOI: 10.1002/msb.201304521.
- Allfrey V.G. & Mirsky A.E. 1964. Structural modifications of histones and their possible role in regulation of RNA synthesis. Science. 144, 559.
- Pazin M.J. & Kadonaga J.T. 1997. What’s up and down with histone deacetylation and transcription? Cell. 89, 325-328.
- Kim S.C. et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 23, 607-618.
- Choudhary C. et al. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325, 834-840.
- Ciechanover A. et al. 2005. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79-87.
- Hay R.T. et al. 2005. SUMO: A history of modification. Mol. Cell. 18, 1-12.
- Minguez P. et al. 2012. PTMcode: A database of known and predicted functional associations between post-translational modifications in proteins. Nucleic Acids Res. 41, D306-D311.
- Lu Z. et al. 2011. Bioinformatic analysis and post-translation modification crosstalk prediction of lysine acetylation. PLoS ONE. 6, e28228.
- Swaney D. el al. 2013. Global analysis of phosphorylation and ubiquitylation crosstalk in protein degradation. Nat. Methods. 10, 676-682.
- Yao Q. et al. 2011. SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J. Biol. Chem. 286, 27342-27349.
- Mertins P. et al. 2013. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods. 10, 634-637.
- Su Y.-F. et al. 2012. Phosphorylation-dependent SUMOylation of the transcription factor NF-E2. PLoS ONE. 7, e44608.
- Hart G.W. et al. 2011. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 525-858.
- Wang Z. et al. 2008. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793-13798.

