Validation of SUMO-2/3 Antibodies (12F3 and 11G2) in Western Blot, Immunoprecipitation and Immunofluorescence Applications
We describe the validation results of newly developed, mouse monoclonal antibodies against SUMO-2/3, 12F3 (Cat # ASM23) and 11G2 (Cat # ASM24). Antibody performance was compared with the popular 8A2 antibody in western blot (WB), immunoprecipitation (IP) and immunofluorescence (IF) applications. The validation data clearly demonstrates that 12F3 and 11G2 have a high sensitivity and specificity against SUMO-2/3. They are able to precipitate SUMO-2/3 targeting proteins efficiently in IP experiments and 12F3 stains SUMO-2/3 substrates strongly in various fixation conditions. Clone 12F3 was shown to outperform the commercially available clone 8A2 in all applications. Clone 11G2 antibody is a poor reagent for Western-Blot application, but an exceptional reagent in IP applications. Our initial data suggests that clones 11G2 and 12F3 immunoprecipitate largely overlapping SUMOylated protein profiles with some differential protein affinity. Clone 11G2 is currently under validation for IF applications. The newly developed anti-SUMO-2/3 antibodies will provide powerful tools for the identification of in vivo and in vitro SUMO-2/3 substrates and their functional roles in cells and tissues.
Introduction
Dynamic post translational modifications (PTMs) provide proteins with vastly expanded functionality. Thus, PTMs have gained a great attention and have been a fast growing research area not only in cellular physiology, but also in pathology. A study of the dynamic events of PTM requires extensively validated antibodies with a high specificity and affinity. Here, we validated newly developed antibodies against SUMO-2/3 in WB, IP and IF applications. SUMO (Small Ubiquitin-like MOdifier) proteins link to target proteins through enzymatic cascades and SUMOylation plays pivotal roles in transcriptional regulation, chromatin remodeling, response to stress and regulation of mitosis1. Here, we present our experimental data for the validation and characterization of two new SUMO-2/3 antibodies.


Results
Western Blot Analysis (WB)
Immunoblot analysis of recombinant SUMO-2 and SUMO-1 demonstrated that 12F3 has a high sensitivity to detect sub-nanograms of SUMO-2/3 without a cross-reactivity to SUMO-1 (more than 1300 fold specificity, Fig 1A). Simple intensity comparison indicates that 12F3 is at least 8 times more sensitive to SUMO-2/3 than the commonly used antibody 8A2 (Fig. 1A), which is corroborated by the immunoblot on total cell lysate (Fig. 1B). Antibody 11G2 was found to be a poor reagent for western blot applications (data not shown).
To further examine antibody specificity, SUMOylation levels were manipulated in a HeLa cell culture system and probed with 12F3 antibody or 8A2 antibody. Cells were treated with 43°C heat shock (Fig 1B, lanes HS43) for 10 min, which is known to enhance SUMO-2/3 SUMOylation2, or subject to SUMO-2 shRNA knock-down (Fig 1B, lanes KD S2). Western blot analysis with total cell lysate demonstrates that both antibodies could identify the changing levels of SUMOylation, 12F3 was more sensitive than 8A2 at the concentration of antibody used in Fig1 (1 µg/ml).
Figure 1: Western Blot Analysis with 12F3 and 8A2
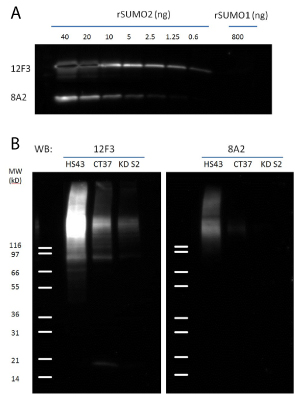
Legend: A) recombinant SUMO-1 and SUMO-2 proteins were detected by 12F3 and 8A2 in a sensitivity and specificity test. 1ug/ml of each antibody was used. B) Three cell lysates were prepared from HeLa cells: Heat Shock treated (HS43, 43°C for 10min), untreated (CT37) and shRNA SUMO-2 knock down (KD S2). 20ug of HeLa cell lysates from each sample were separated by SDS-PAGE and probed with 12F3 and 8A2. Note that the expression of SUMO-3 is not affected by SUMO-2 knock down. Signal in the lane KD S2 is likely from SUMO-3.
Immunoprecipitation Analysis (IP)
12F3 and 11G2 antibodies were used to immunoprecipitate SUMO-2/3-proteins in HeLa cell lysates. Immunoprecipitation efficiency was compared to that of the commercially available clone 8A2. Endogenous SUMO-2/3 SUMOylated proteins were enriched and total precipitates and a specific target protein, TFII-I were probed.
Cell lysates from heat shocked, untreated and SUMO-2 knockdown HeLa cells were used for immunoprecipitation analysis (Fig. 2A). In all cases more proteins were enriched by both 12F3 and 11G2 than by 8A2, suggesting that these newly developed antibodies will be powerful reagents in the characterization of protein SUMOylation.
It was noted that 11G2 precipitates free SUMO-2/3 more efficiently than 12F3 (Fig. 2A), which may imply that these two antibodies have different preference to SUMO-2/3 conjugates. Distinctive selectivity for SUMO-2/3 targets by clones 12F3 and 11G2 is also suggested by the fact that TFII-I (General transcription factor II-I) enrichment is stronger with 11G2 than 12F3 (Fig. 2B). Notably, 8A3 is a much weaker IP reagent than either 12F3 or 11G2. Only when 3-5 fold higher amount of 8A2 is used3, TFII-1 enrichment can be observed (data not shown). Note that in Input lane (IP) unconjugated TFII-I is visible near 120kDa. Multiple bands indicate that TFII-I is SUMOylated by several SUMO-2/3 proteins, as has been reported previously4.
Figure 2: Immunoprecipitation with 12F3, 11G2 and 8A2
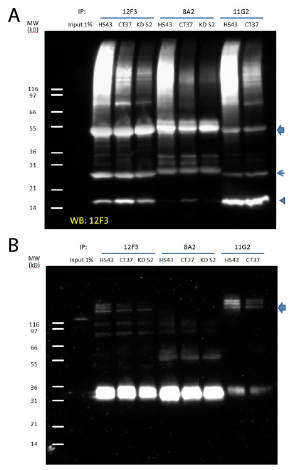
Legend: Denatured cell lysates were prepared from HS43, CT37 and KD S2. 1mg of lysate was used for the immunoprecipitation of SUMO-2/3 modified proteins. Cell lysates were incubated with antibodies (30 µl at 1 µg/µl), and then bound to protein G beads (30ul slurry). Western blots of immunoprecipitated proteins were developed using 12F3 (A) or anti-TFII-I antibody (B). Arrow head indicates free SUMO-2/3. Arrows indicate heavy and light chains of antibodies.
Immunofluorescence Analysis (IF)
SUMO-2/3 conjugates in fixed HeLa cells were visualized by using 12F3 antibody (2ug/ml, Fig 3A). Cells were permeabilized with 40ug/ml digitonin solution and fixed with 4% paraformaldehyde. Mitotic cells in metaphase were selected and the co-localization of microtubules, SUMO-2/3 and chromosomes was examined by confocal microscopy. SUMO-2/3 conjugated proteins exist mainly as puncta throughout cell body, but are heavily localized on chromosomes during mitosis, as reported previously5. In contrast to 12F3 signal from 8A2 was much weaker at the same concentration (2ug/ml, Fig 3B). When higher concentration (20ug/ml) of 8A2 was used, puncta on chromosome were visible, but still their intensity was not comparable to 2ug/ml of 12F3 (Data not shown).
As reported previously, we required a digitionin permeabilization step to clearly visualize SUMO-2/3 localization to chromosomes in mitotic cells6. Staining in paraformaldehyde-Triton X-100 fixation gave good signals for non-mitotic cells (data not shown).
Figure 3: Immunofluorescent staining with 12F3 and 8A2
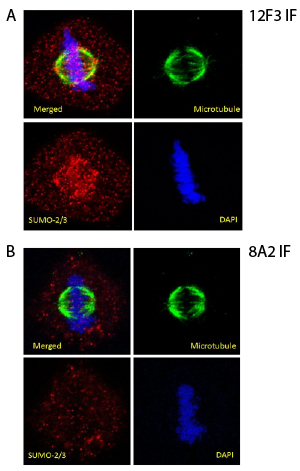
Legend: HeLa cells were permeabilized with digitonin solution (40µM for 1 min) and fixed in 4% paraformaldehyde (10min). The cells were stained against β-tubulin (green) and SUMO-2/3 12F3 (Fig. 3A) or 8A2 (Fig. 3B). DNA was stained with DAPI. Mitotic cells in metaphase were imaged with a Zeiss LSM 780 confocal microscope (1.4 NA 63X objective).
Discussion
A critical step in PTM studies is the identification of the target proteins for a given PTM, which mainly relies on good antibodies. The enrichment of target proteins by immunopreciptation is routinely carried out to identify the PTM targets, usually going through the downstream process of mass spectrometry7. This approach is often limited by the lack of high quality antibodies of high specificity and sensitivity. Low specificity of an antibody will introduce contaminant proteins in the analysis and give false positive and poor sensitivity will not bring target proteins into even consideration. In order to avoid these problems, many scientists employ overexpression of protein mutants containing a tag (e.g. HA and Flag), against which antibodies of high quality are commercially available. However, this mutant expression is time consuming and labor intensive, often disturbs a well-balanced cell system and does not work in many cell lines. Newly developed antibodies (12F3 and 11G2) against SUMO-2/3 are powerful new tools for the study of SUMOylation of endogenous proteins. Their binding affinity is strong enough to precipitate SUMO-2/3 conjugates very efficiently. Clone 12F3 shows superior performance in various applications including WB, IP and IF (ChIP application is under validation). While 11G2 does not appear to be a good choice for western blot applications, it has been shown to be an excellent reagent for IP applications and it’s IF application is a under validation (Table 1). We expect that these two antibodies will enable scientists to perform a convenient and straight-forward experiments and will help to expand our knowledge of SUMOylation.
Table 1: Summary of Antibody Comparisons

Materials and Methods
Reagents
Monoclonal mouse antibodies (12F3 and 11G2) were developed and manufactured at Cytoskeleton Inc. (cat.# ASM23 and ASM24, respectively) and 8A2 antibody was purchased from Abcam (cat.# ab81371). HeLa cells (cat# CCL-2) were purchased from ATCC. Protease inhibitor cocktail (cat# PIC02) and sheep anti-tubulin antibody (cat#ATN02) were from Cytoskeleton Inc. Alexa-Fluor 488 donkey anti-sheep antibody (cat# 713-545-147) and Alexa-Fluor 555 donkey anti-mouse antibody (cat# A31570) were purchased from Life Technologies. HRP conjugated goat anti-mouse IgG (cat# 115-035-068) was from Jackson Immunoresearch Labs. Protein G-beads (cat# 6511-100) were from BioVision. Digitonin (cat# D141) and NEM (cat# E3876) were purchased from Sigma.
Materials
Glass bottom dish (cat# P35G-1.5-14-C) was purchased from MatTeck.
Western blot method
Samples were run on SDS-PAGE using 4-20% Tris-Glycine gels. Gels were equilibrated in western blot buffer (25 mM Tris pH8.8, 192 mM glycine, 15% methanol) for 15 min. at room temp. prior to electroblotting. The separated proteins were transferred to a PVDF membrane overnight at constant 20V and 4°C. PVDF was washed once in TBST for 10 min. and blocked in 5%milk in TBST for 60 min. at room temp. with constant agitation. The membrane was then incubated with a 1:1000 dilution of primary antibody in TBST at room temp. for 1h with constant agitation. Membranes were washed three times in TBST for 10 min. each at room temp. then incubated with a 1:20,000 dilution of HRP-conjugated goat anti-mouse IgG in TBST for 1h. at room temp. with constant agitation. The membrane was washed 4 times in TBST for 10 min. each and HRP signal was detected by chemilluminescence.
Immunoprecipitations
HeLa cell lysate was prepared as described previously (3) 30ug of antibody was incubated with 1mg cell lysate for 1hr in ice. 30 ul of Protein G slurry was added and incubated for 2h at 4°C with a rotation. Protein G beads were washed 3 times by a centrifugation (960 x g, 4°C, 1 min) and resuspended with washing buffer (50mM Tris pH7.5, 150mM NaCl, 1% IGEPAL). Captured proteins were released from the beads by resuspending Protein G pellets in 30 µl of 2X non-reducing SDS sample buffer (125mM Tris pH6.8, 20% glycerol, 4% SDS, 0.005% Bromophenol blue). The solution was incubated at room temperature for 5 min and a supernatant (~30ul) was collected after centrifugation (960 x g, 1 min., room temp). The supernatant was boiled with 1ul of beta mercaptoethanol for 5 min. The sample was analyzed with SDS-PAGE and immunoblotting.
IF method
HeLa cells were grown on a glass bottom dish to 70~80% confluence. Cells were permeabilized by incubating with digitonin solution (20mM HEPES pH 7.4, 110mM Potassium acetate, 2mM Magnesium acetate, protease inhibitor cocktail, 10mM NEM and 40ug/mL digitonin) for 1 min with gentle agitation, followed by 4% paraformaldehyde fixation (10 min)6. Cells were incubated with 500 µl of primary antibodies (1:500 dilution in PBS) at room temp. for 45 min. The sample was washed two times (10min. each wash) in PBS (3ml per wash). Secondary antibodies at 1:500 dilution plus DAPI at 100ng/ml were placed on the cells and incubated at room temp. for 45 min. Samples were washed with PBS as described above and cells were observed by Zeiss LSM 780 confocal microscope (1.4 NA 63X objective).
References
1. Willson V. et al. 2009. SUMO Regulation of Cellular Processes. Springer.
2. Saitoh H. & Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252-6258.
3. Barysch et al. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 2014 Apr;9(4):896-909
4. Becker et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013 Apr;20(4):525-31.
5. Ayaydin F. & Dasso M. 2004. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 15, 5208-5218.
6. Zhang et al. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008 Mar 28;29(6):729-41
7. Alfred et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics.Mol Cell Proteomics. 2006 Dec 5(12):2298-310
Related Links
Anti-SUMO 2/3 Antibody Clone 12F3 (Cat. # ASM23)
Anti-SUMO 2/3 Antibody Clone 11G2 (Cat. # ASM24)
Anti-Phosphotyrosine Mouse Monoclonal Antibody Clone 27B10.4 (Cat. # APY03)
Anti-Acetyl Lysine Monoclonal Antibody (Cat. # AAC01)
Anti-Ubiquitin Monoclonal Antibody (Cat. # AUB01)
