Investigating PTMs: Beyond Prediction and Identification
Summary of PTMs
Post-translational modifications (PTMs) are dynamic, tightly-regulated alterations to a protein that control the protein’s structure, spatial localization, and interactions; thereby, governing its function [1-3]. See references for in-depth reviews.
With the advancement in PTM identification, championed by the mass spectrometry [4-5] and computational biology fields [6-8], scientists now estimate that a million or more modified proteoforms exist in mammalian cells [9-10]. With this plethora of information available, do we need molecular and cell biologists studying PTMs? Yes, because while remarkable progress has been made in growing the PTM proteome, on its own provides little insight towards defining a PTM’s role on a target protein’s function, activity, or biological outcome.
Why is understanding PTM function so important? PTM modified proteins have been shown to regulate critical biological processes, many of which are dysregulated in disease [11-15]. Scientist’s understanding of a few PTMs on a select number of target proteins has even led to therapeutic drug targets [16-18]; still, there are hundreds of thousands more proteoforms out there waiting to be investigated with in-depth functional studies.
Large proteomic data sets provide a strong foundation for hypothesis development, but are just the tip-of-the-iceberg; ultimately, mechanistic investigation of target proteins PTMs are necessary for a complete understanding of their importance physiologically and pathologically. Here we provide tips that will ensure optimal approaches are used when performing functional, biological studies on target protein PTMs.
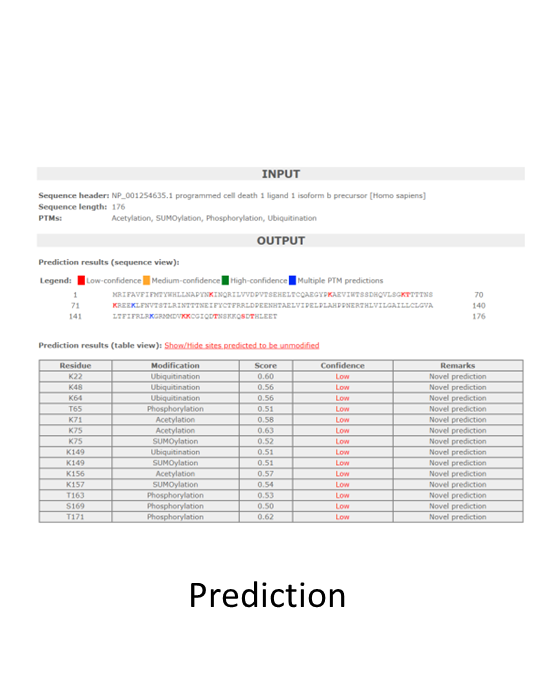
Mass spectrometry (MS)-proteomics in combination with the plethora of PTM prediction tools now available make it quite simple to gain insight into whether or not a protein may be modified by a particular PTM. For more information about mass spectrometry based PTM proteomics click here. Once a potentially interesting PTM for a target protein has been identified by these approaches, what’s the next step?
For cell and molecular biologists there is a critical step that occurs when taking a target identified from informatics data and investigating it in their biological system. This essential step is validation. Why is it so critical? In-depth investigation of an unexplored PTM of a target protein hinges on whether or not the modified target can be effectively identified in the investigator’s biological system. Utilizing sub-optimal approaches may result in the inability to detect the modified protein; thus, leading to a premature end to potentially novel discoveries.
As an example, the PTM, ubiquitin, was first shown to interact with microtubule networks several decades ago in 1988 [19]. However, at that time the data suggested that ubiquitin was modifying microtubule-associated proteins but not tubulin itself [19], which may have played a role in the nearly two decade dormancy in the ubiquitinated tubulin field. Unfortunately, these findings were based on western blotting data performed without any enrichment techniques [19]. With the advancement in PTM detection techniques, there are now several papers that have successfully identified ubiquitinated tubulin; furthermore, these studies link tubulin ubiquitination with microtubule dynamics and function [20-23]. To learn more about the importance of tubulin ubiquitination see our recent newsletter. This example highlights how vital it is to use optimal approaches and tools when validating whether or not a protein is post-translationally modified.
Our PTM Detection Techniques page provides a summary of enrichment techniques and validation approaches like PTM IP, protein IP, and PTM and/or protein overexpression. Furthermore, Cytoskeleton offers comprehensive PTM detection kits that have been optimized for maximal detection of endogenous PTMs [24]. A recent publication utilized these kits to examine the endogenous PTM states of proteins in the EGFR-Ras-c-Fos axis [24].
Points to Consider When Investigating Functional Importance of Target Protein PTMs
The following sections will discuss critical points to consider when investigating PTMs such as dealing with transient changes, the importance of studying PTMs endogenously, how to minimize false negative and positive results, as well as, the need to consider investigating PTM crosstalk.
For technical tips on studying target protein PTMs using enrichment strategies click here.
Want to see a peer-reviewed video protocol [25] for endogenous PTM enrichment and detection? Click here.
Studying transient and dynamic PTM changes
An overarching problem with studying PTMs is their transient nature which often leads to low stoichiometry (< 1-5%) [26-28]. While an impediment towards their investigation, these qualities also make them key regulatory agents or switches for rapid biological functions in cells.
A common approach to overcome this barrier is to enrich for the PTM of interest for a target protein. Enrichment can be achieved by two paths: 1. Enrich for the PTM using affinity matrices and probing for the protein of interest or 2. Enrich for the protein using affinity matrices and probing for the PTM. Click here for specific tips for success when performing these enrichment strategies. We recommend a combination of these approaches to properly validate that a protein is modified by a target PTM; however, as a first approach enriching for the PTM may be a more successful strategy.
The robust Signal-Seeker tools can detection PTMs as low as 17 molecules per cell [24]. The ability to detect these small, but potentially meaningful changes are critical, as measuring endogenous changes of PTMs are necessary to understand the physiologic relevance of the modification as discussed below.
Investigate PTMs endogenously and physiologically
Another approach to circumvent low levels of PTM modified proteins is to exogenously overexpress a PTM, target protein, and in many cases both. This approach will provide insight into whether or not a target protein is modified by a PTM of interest; however, there are cases where false interactions between an overexpressed protein or PTM may occur [29].
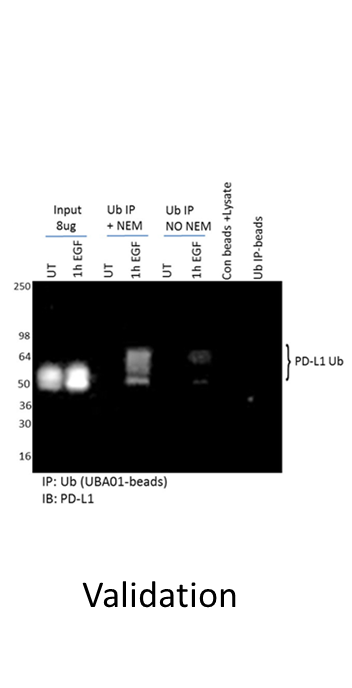
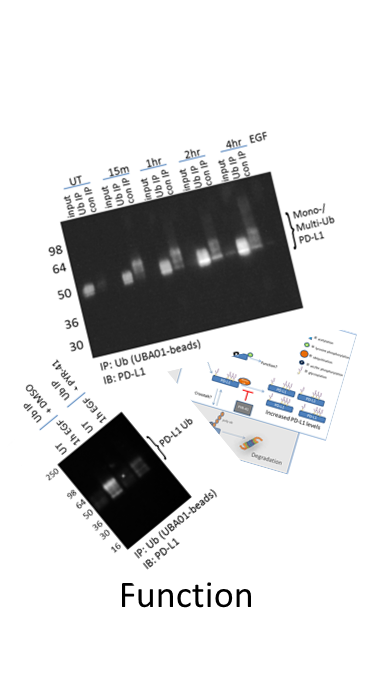
Another problem with overexpression systems is that they are often used for follow-up physiologic studies of the modified target. For example, a study investigating Ras di-ubiquitination (di-Ub) using overexpression systems determined that the modification, while important for Ras activation, was not regulated by upstream EGFR signaling [30]. Contrarily, endogenous Ras di-Ub was examined using Cytoskeleton’s Ubiquitin Detection Kit, and robust, dynamic changes in di-Ub of Ras occurred in response to EGF stimulation [24]. It is likely that the overexpression system masked physiologic changes in di-Ub Ras. These data highlight the importance of understanding the limitations of specific tools, as well as identifying robust tools that effectively capture endogenous, physiologic changes in a target protein’s PTM profile.
The Signal-Seeker PTM Detection toolkits have been used to effectively measure the endogenous level of multiple PTMs for many target proteins [24,31,32], and all of these studies can be found on our validation data page.
Minimize false positive, false negative data
Negative detection of a PTM for a target protein can be due to a multitude of reasons both biologically and technically; therefore, a negative result does not definitively prove that the protein is incapable of being modified. For example, a particular target protein may only be modified under specific biological conditions in a specific cell type, which may not have been examined in the available literature. Often it is only a very limited pool of researchers that have studied any given protein of interest (POI), and therefore have the expertise and insight to know what experimental system, conditions, and timelines are necessary to study their POI. A set of tools that empower these researchers to investigate their proteins PTM state would greatly facilitate PTM studies.
Another factor that may produce false positive or false negative data is protein–protein interactions that are not disrupted during cell lysis. Therefore, it is recommended to utilize stringent buffers that are compatible with the affinity reagent. Cytoskeleton has developed a unique denaturing lysis buffer system that is compatible with all of our PTM detection systems to minimize the potential for protein-protein interference.
As with all enrichment techniques that utilize affinity matrices the possibility for non-specific binding to PTM affinity matrices is a possibility that may produce false positive data. Thus, specific IgG control beads will help to delineate specific from non-specific interactions. Cytoskeleton offers matrix-matched control beads for all of our PTM affinity matrices as having the right controls will simplify data interpretation.
It is noteworthy to consider the potential for false negative detection of endogenous PTM changes with IP-pan-PTM methodologies. For example, the protein dihydrolipoamide dehydrogenase (DLD) is acetylated at potentially 22 different lysine residues. Thus, one prominent Ac site on DLD may mask small, but meaningful changes that occur at alternative lysine residues on the protein. This hypothetical example highlights the importance of investigating PTMs using multiple approaches, as site-directed mutagenesis would be beneficial for these types of analysis. Additionally, it further enhances the call for additional PTM tools that enable site specific investigation; for example, site specific PTM antibodies.
Approaches to investigate PTM crosstalk
Common crosstalk paradigms have emerged [33,34], such as phosphorylation being a precursor to target protein Ub [35,36]. Another pattern of crosstalk exists between Ac and Ub where these modifications compete to regulate the stability of the protein [37,38]. Ac has been shown to stabilize target proteins [39], while poly-Ub is well characterized as a mark for proteasomal degradation [40]. Both Ac and Ub primarily modify lysine residues, and many proteins have been identified to be both acetylated and ubiquitinated on the same lysine residue, providing further evidence of negative crosstalk between these PTMs.
Signal-Seeker PTM detection tools have the ability to investigate multiple PTMs of interest, which allows investigators to gain insight into potential PTM crosstalk. This is possible because all of the affinity matrices used to capture the PTMs of interest were designed to work with a universal lysis buffer as reported previously [25], which enables more meaningful comparisons when investigating crosstalk between PTMs.
For more specific information about Signal-Seeker™ post-translational modification detection tools please view our product pages, resource links, or contact technical support.
Additional Links:
How to study PTMs: A Guide for new PTM Investigators
Post-Translational Modification Detection Techniques
Bioscience Reports Article: Simple toolsets to identify endogenou PTMs for a target protein
Neoplasia Article: Identifying Regulatory Posttranslational Modifications of PD-L1
JoVE Article: Visual Protocol For Investigating Post- translational Modifications of Target Proteins
Monoubiquitination: A dynamic tag for protein regulation
References
1. Seo J, Lee KJ. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37(1):35-44,2. Bah A, Forman-Kay JD. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J Biol Chem. 2016;291(13):6696-705, 10.1074/jbc.R115.695056.3. Buuh ZY, Lyu Z, Wang RE. Interrogating the Roles of Post-Translational Modifications of Non-Histone Proteins. Journal of medicinal chemistry. 2017, 10.1021/acs.jmedchem.6b01817.4. Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12(12):3444-52, 10.1074/mcp.O113.034181.5. Doll S, Burlingame AL. Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem Biol. 2015;10(1):63-71, 10.1021/cb500904b.6. Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150(2):413-25, 10.1016/j.cell.2012.05.036.7. Li S, Iakoucheva LM, Mooney SD, Radivojac P. Loss of post-translational modification sites in disease. Pac Symp Biocomput. 2010:337-47,8. Nickchi P, Jafari M, Kalantari S. PEIMAN 1.0: Post-translational modification Enrichment, Integration and Matching ANalysis. Database (Oxford). 2015;2015:bav037, 10.1093/database/bav037.9. Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8(1):33-41, 10.1016/j.cbpa.2003.12.009.10. Regnier FE, Kim J. Proteins and Proteoforms: New Separation Challenges. Anal Chem. 2018;90(1):361-73, 10.1021/acs.analchem.7b05007.11. Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19(1):59-70, 10.1038/nrm.2017.83.12. Basak S, Lu C, Basak A. Post-Translational Protein Modifications of Rare and Unconventional Types: Implications in Functions and Diseases. Curr Med Chem. 2016;23(7):714-45,13. Anbalagan M, Huderson B, Murphy L, Rowan BG. Post-translational modifications of nuclear receptors and human disease. Nucl Recept Signal. 2012;10:e001, 10.1621/nrs.10001.14. Liddy KA, White MY, Cordwell SJ. Functional decorations: post-translational modifications and heart disease delineated by targeted proteomics. Genome Med. 2013;5(2):20, 10.1186/gm424.15. Droescher M, Chaugule VK, Pichler A. SUMO rules: regulatory concepts and their implication in neurologic functions. Neuromolecular Med. 2013;15(4):639-60, 10.1007/s12017-013-8258-6.16. Su MG, Weng JT, Hsu JB, Huang KY, Chi YH, Lee TY. Investigation and identification of functional post-translational modification sites associated with drug binding and protein-protein interactions. BMC Syst Biol. 2017;11(Suppl 7):132, 10.1186/s12918-017-0506-1.17. West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30-9, 10.1172/JCI69738.18. Ono M, Kuwano M. Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to and other EGFR-targeting drugs. Clin Cancer Res. 2006;12(24):7242-51, 10.1158/1078-0432.CCR-06-0646.19. Murti KG, Smith HT, Fried VA. Ubiquitin is a component of the microtubule network. Proc Natl Acad Sci U S A. 1988;85(9):3019-23,20. Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28(8):868-73, 10.1038/nbt.1654.21. Kohta R, Kotake Y, Hosoya T, Hiramatsu T, Otsubo Y, Koyama H, et al. 1-Benzyl-1,2,3,4-tetrahydroisoquinoline binds with tubulin beta, a substrate of parkin, and reduces its polyubiquitination. J Neurochem. 2010;114(5):1291-301, 10.1111/j.1471-4159.2010.06576.x.22. Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell Cycle. 2010;9(5):980-94, 10.4161/cc.9.5.10934.23. Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci. 2003;23(8):3316-24,24. Horita H, Law A, Hong S, Middleton K. A simple toolset to identify endogenous post-translational modifications for a target protein: a snapshot of the EGFR signaling pathway. Biosci Rep. 2017, 10.1042/BSR20170919.25. Horita H, Law A, Middleton K. Utilizing a Comprehensive Immunoprecipitation Enrichment System to Identify an Endogenous Post-translational Modification Profile for Target Proteins. J Vis Exp. 2018(131), 10.3791/56912.26. Johnson H, Eyers CE. Analysis of post-translational modifications by LC-MS/MS. Methods Mol Biol. 2010;658:93-108, 10.1007/978-1-60761-780-8_5.27. Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, et al. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 2011;8(8):677-83, 10.1038/nmeth.1636.28. Ordureau A, Munch C, Harper JW. Quantifying ubiquitin signaling. Mol Cell. 2015;58(4):660-76, 10.1016/j.molcel.2015.02.020.29. Emmerich CH, Cohen P. Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem Biophys Res Commun. 2015;466(1):1-14, 10.1016/j.bbrc.2015.08.109.30. Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21(5):679-87, 10.1016/j.molcel.2006.02.011.31. Horita H, Law A, Middleton K. Utilizing Optimized Tools to Investigate PTM Crosstalk: Identifying Potential PTM Crosstalk of Acetylated Mitochondrial Proteins. Proteomes. 2018;6(2), 10.3390/proteomes6020024.32. Horita H, Law A, Hong S, Middleton K. Identifying Regulatory Posttranslational Modifications of PD-L1: A Focus on Monoubiquitinaton. Neoplasia. 2017;19(4):346-53, 10.1016/j.neo.2017.02.006.33. Csizmok V, Forman-Kay JD. Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications. Current opinion in structural biology. 2018;48:58-67, 10.1016/j.sbi.2017.10.013.34. Lothrop AP, Torres MP, Fuchs SM. Deciphering post-translational modification codes. FEBS Lett. 2013;587(8):1247-57, 10.1016/j.febslet.2013.01.047.35. Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10(7):676-82, 10.1038/nmeth.2519.36. Filipcik P, Curry JR, Mace PD. When Worlds Collide-Mechanisms at the Interface between Phosphorylation and Ubiquitination. J Mol Biol. 2017;429(8):1097-113, 10.1016/j.jmb.2017.02.011.37. Ryder DJ, Judge SM, Beharry AW, Farnsworth CL, Silva JC, Judge AR. Identification of the Acetylation and Ubiquitin-Modified Proteome during the Progression of Skeletal Muscle Atrophy. PLoS One. 2015;10(8):e0136247, 10.1371/journal.pone.0136247.38. Butler PL, Staruschenko A, Snyder PM. Acetylation stimulates the epithelial sodium channel by reducing its ubiquitination and degradation. J Biol Chem. 2015;290(20):12497-503, 10.1074/jbc.M114.635540.39. Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23(7):2587-99,40. Wagner SA, Beli P, Weinert BT, Scholz C, Kelstrup CD, Young C, et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 2012;11(12):1578-85, 10.1074/mcp.M112.017905.