Introduction
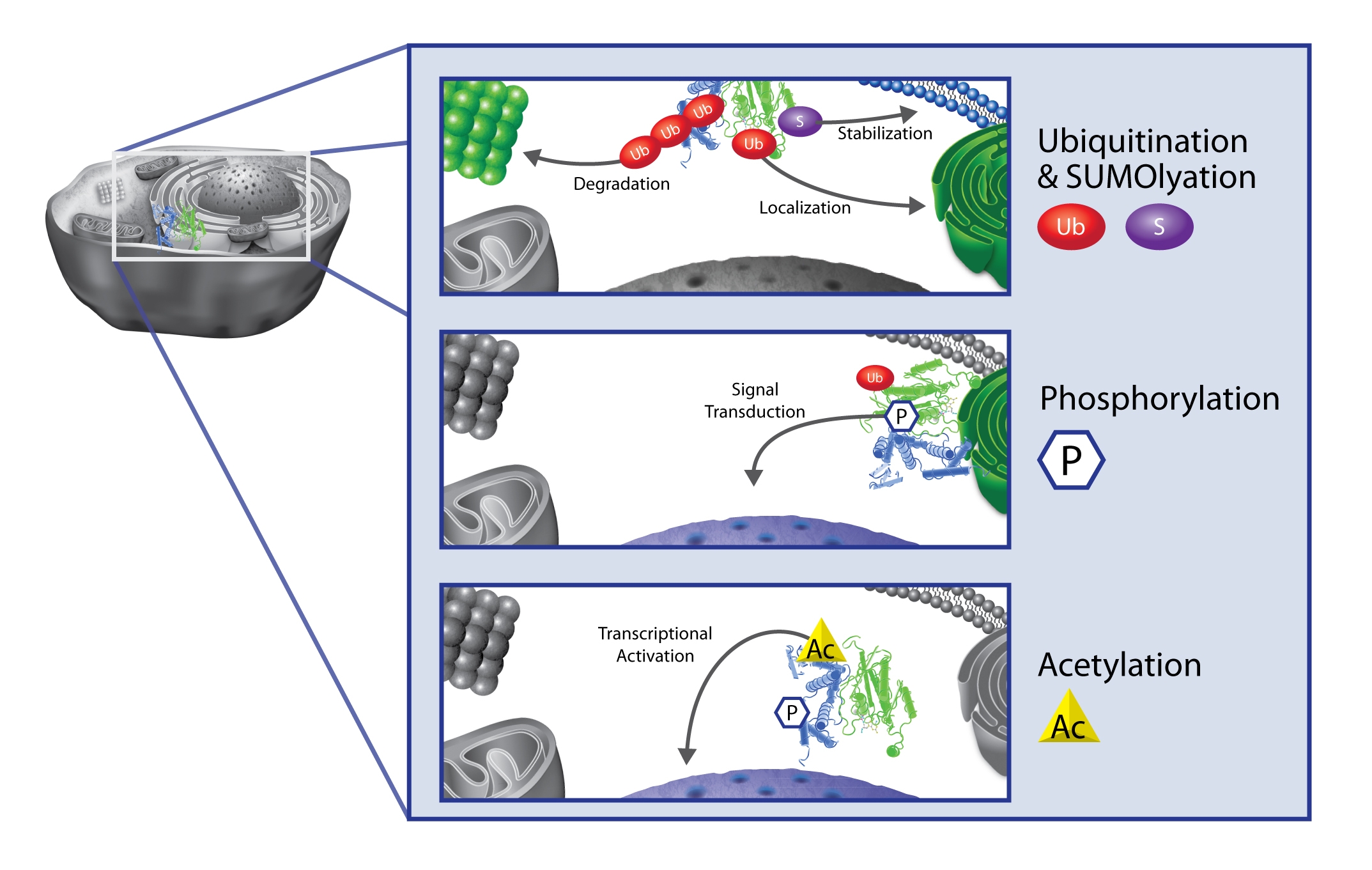
The mammalian proteome has been estimated to contain multiple millions of unique proteoforms. This level of complexity is derived from a relatively simple genome (approx. 25,000 genes), a transcriptome which increases the potential protein footprint to 100,000, and protein post-translational modifications (PTMs) which account for the vast increase in proteome complexity and an almost limitless potential for functional diversity. This information suggests that nearly all proteins will be modified by multiple PTMs. For any given protein, a variety of PTM proteoforms offer a way to facilitate rapid cellular changes by altering the structure and function of the protein.
In many cases PTMs have been shown to work in concert to orchestrate a specific protein function and recent studies have suggested that both cooperative and negative PTM crosstalk is a pervasive and fundamental cell regulatory mechanism.
Importantly for human health and disease, misregulation of PTMs has been implicated in the progression of diseases like cancer, heart failure, neurologic, and metabolic diseases.
See the table below for information about key, regulatory protein modifications, or go to ModPred (also found in our resources link) to see if your target protein may potentially be modified.
PTM | Established Function | Novel Function | Signal Transduction | Target Amino Acid(s) | PTM Crosstalk | Modifiers |
epigenetic and metabloic regulation | protein stability, localization, synthesis | Yes | K | Yes | HAT, HDAC | |
epigenetic regulation | protein: protein interaction, signal transduction | Yes | R,K | Yes | methyltransferase, demethylase | |
protein folding, structure, and stability | cell adhesion, trafficking of glycoproteins | Yes | N | Yes | glycosyltransferase, glycoside hydrolase | |
protein folding, structure, and stability | may compete with phosphorylatin for S,T binding | Yes | S,T | Yes | glycosyltransferase, glycoside hydrolase | |
signal transduction | protein:protein interaction, precursor to degradation, epigentics | Yes | Y, S, T | Yes | Kinase, phosphatase | |
membrane protein association | subcellular trafficking of proteins, protein:protein interaction, stability | dynamic reversible modification | C, S, T | Yes | palmitoyl protein thioesterases | |
nuclear protein localization, transcriptional activity | protein stability, response to stress, cell cycle | Yes | K | Yes | SAE, ubc9, SENP | |
proteasomal targeting - degradation | protein localization, activity, protein:protein interaction | Yes | K | Yes | Ub Ligase, DUB |
a
Additional Links:
Overview of PTM Detection Techniques and Methods
a
Cytoskeleton is committed to providing the most comprehensive reagents that allow highly sensitive and specific detection of endogenous protein modifications. Signal-Seeker™ kits use affinity beads to isolate and enrich modified proteins from any given cell or tissue lysate. Quality affinity reagents are key to allowing enrichment of often very transient and low level modified species, the affinity beads developed by our scientist are highly optimized to maximie the probability of identifying key regulatory modifications of even the rarest species.
Advantages
• Unparalleled sensitivity due to optimized kit reagents
• Discover and publish novel regulatory mechanisms
• Confirm transfection & proteomic results with endogenous data
• Examine protein modification crosstalk