Arp2/3 protein complex: porcine brain
Product Uses Include
- Positive control for in vitro actin polymerization stimualting compounds
- Discovery of Arp2/3 activating or inhibiting compound
- Characterization and discovery of Arp2/3 interacting proteins
Material
The Arp2/3 protein complex consists of 7 protein subunits, all present in approximately equal stoichiometry (see Figure 1). The Arp2/3 complex is a key regulator of actin filament nucleation and the protein subunits are highly evolutionarily conserved. The protein complex has been purified from porcine brain and is supplied as a lyophilized powder. When reconstituted with distilled water, the complex is in the following buffer: 20 mM Tris pH 7.5, 25 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM ATP, 1% dextran and 5% sucrose. The molecular weight of the Arp2/3 complex is 224 kDa.
Note: RP01P from procine brain replaces RP01 from bovine brain. We have compared the activity of Arp2/3 complex from both sources and have found them to behave identically in actin polymerization assays (see Fig. 2)
Purity
Protein purity is determined by scanning densitometry of Coomassie Blue stained protein on a 4-20% polyacrylamide gel. Arp2/3 protein complex is determined to be 90% pure (see Figure 1).
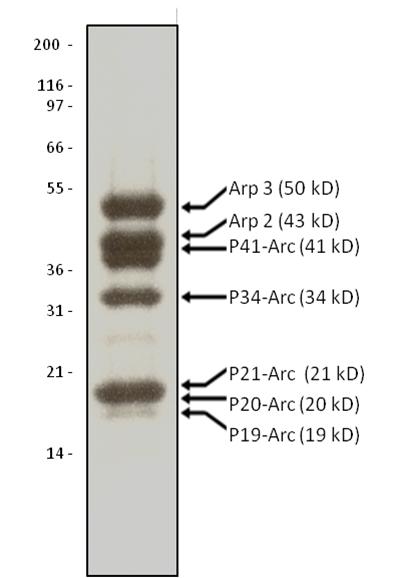
Figure 1. Arp2/3 Protein Purity Determiniation: A 10 μg sample of Arp2/3 complex was separated by electro-phoresis in a 4-20% SDS-PAGE system and stained with Coomassie Blue. Protein quantita-tion was determined with the Precision RedTM Protein Assay Reagent (Cat. # ADV02). Mark12 molecular weight markers are from Life Technologies.
Biological Activity
RP01P was tested in an actin polymerization assay (Cat. # BK003). In conjunction with the VCA domain of WASP (Cat. # VCG03), an Arp2/3 activator, it was shown to stimulate actin polymerization by 20-fold compared to the control without RP01. This indicates Arp 2/3 specificity for actin polymerization induction (Figure 2).
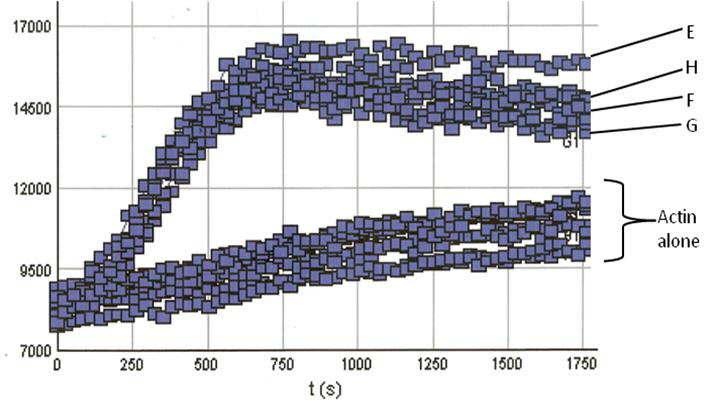
Figure 2. Actin Polymerization Assay with Arp2/3 and VCA domain proteins: Actin polymerization was carried out as described in the method; All reactions contain 0.8 μM actin. Reactions containing Arp2/3 or VCA (data not shown) show little or no enhancement of actin nucleation. Actin alone also shows low polymerization com-petence under these conditions (Actin alone reactions). In the presence of the VCA domain however, Arp2/3 results in an en-hancement of actin nucleation (Note the steep nucleation phase of polymerization). Actin polymerization is measured in arbitrary fluorescent units over time. In our tests porcine derived Arp2/3(wells E & F) behaved identically to Arp2/3 derived from bovine sources (wells G & H).
For product Datasheets and MSDSs please click on the PDF links below. For additional information, click on the FAQs tab above or contact our Technical Support department at tservice@cytoskeleton.com
Note: RP01P from procine brain replaces RP01 from bovine brain. We have compared the activity of Arp2/3 complex from both sources and have found them to behave identically in actin polymerization assays. The assays below reference the Arp2/3 protein complex: bovine brain (Cat. RP01). Please inquire to tservice@cytoskeleton.com for the latest papers citing this product.
Leng, Y., Zhang, J., Badour, K., Arpaia, E., Freeman, S., Cheung, P., Siu, M. and Siminovitch, K. (2005). Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl. Acad. Sci. U. S. A. 102, 1098-1103.
van der Gucht, J., Paluch, E., Plastino, J. and Sykes, C. (2005). Stress release drives symmetry breaking for actin-based movement. Proc. Natl. Acad. Sci. U. S. A. 102, 7847-7852.
Question 1: What is the best way to store Arp2/3 (Cat. # RP01P)?
Answer 1: Store the lyophilized Arp2/3 protein desiccated (<10% humidity) at 4°C where it is stable for 6 months. Lyophilized protein can also be stored desiccated at -70°C where it will be stable for 6 months. However, at -70°C the rubber seal in the lid of the tube could crack and allow in moisture. Therefore we recommend storing at 4°C. If stored at -70°C, it is imperative to include desiccant with the lyophilized protein if this storage condition is utilized. Resuspend the protein complex to 5 mg/ml with 10 μl of cold Milli-Q water. When resuspended, the complex is in the following buffer: 20 mM Tris pH 7.5, 25 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM ATP, 1.0% (v/v) dextran and 5% (v/v) sucrose. The protein should then be aliquoted into experiment-sized amounts, snap frozen in liquid nitrogen and stored at -70°C where it is stable for 6 months. Further dilution of Arp2/3 should be made in the following buffer: 20 mM Tris pH 7.5, 25 mM KCl, 1 mM MgCl2 and 1 mM DTT (Note: add DTT to the buffer immediately prior to use). NOTE: It is very important to snap freeze the Arp2/3 protein in liquid nitrogen as other methods of freezing will result in significantly reduced activity. Defrost rapidly by placing in a room temperature water bath for 1 min. Avoid repeated freeze/thaw cycles.
Question 2: Is the Arp2/3 (Cat. # RP01P) compatible with the actin polymerization kit (Cat. # BK003)?
Answer 2: Yes, Arp2/3 works very well with the actin polyermization kit (Cat. # BK003) to study the effects of this actin binding protein on actin polymerization. Please see the Arp2/3 datasheet (Cat. # RP01P) for additional information. The Arp2/3 complex is able to induce the branched polymerization of actin filaments in vitro at a molar ratio of 1:200 (Arp2/3:actin). This stimulation is observable in an in vitro polymerization assay; however, the stimulation from Arp2/3 alone is very low under typical polymerization conditions. In the presence of N-WASP protein (or the VCA domain of N-WASP, Cat. # VCG03), the nucleating activity of Arp2/3 is greatly enhanced. In the polymerization assay described in the Arp2/3 datasheet, pyrene actin (Cat. # AP05 or BK003) is present at a final concentration of 0.8 μM, the Arp2/3 complex is at 10 nM and the VCA domain (Cat. # VCG03) is at 400 nM.
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com.





