August 2019 Newsletter : Rac1B, Cancer, and Rac1
 |
| View PDF Here |
The Rac1B GTPase is an alternative splice variant of Rac1, and both GTPases are members of the Rho sub-family of the Ras super-family of GTPases. Insertion of 19 additional amino acids (a.k.a. exon 3b) in Rac1B confers faster GEF-independent GDP/GTP nucleotide exchange due to reduced affinity for GDP and reduced intrinsic GTP hydrolysis compared to Rac11-3(Fig. 1). Biochemically, Rac1B behaves similarly to a constitutively-active Rac1 GTPase, but through a mechanism distinct from oncogenic Rho sub-family GTPase mutants4. Rac1B participates in many phases of oncogenesis, including regulation of cell cycle progression and increased resistance to apoptosis. Additionally, the GTPase has multiple roles in tumor progression, including malignant transformation, epithelial-mesenchymal transition (EMT), metastasis, and invasion3. This newsletter briefly discusses some of the more recent findings regarding the role of Rac1B in tumorigenesis and its relationship with Rac1.
Rac1B is widely regarded as a pro-tumorigenic GTPase as studies find that Rac1B promotes cellular transformation in NIH3T3 mouse fibroblasts and may enhance tumor progression in cancers that over-express Rac1B (e.g., colorectal cancer, human lung adenocarcinomas, thyroid carcinomas)3,5. Rac1B’s role in tumorigenesis involves activation of pathways that result in chronic inflammation, transcription of pro-proliferative genes, inhibition of signaling pathways which inhibit growth and stimulate cell cycle arrest, and modulation of signaling pathways that reduce cell adhesion and promote cell migration3(Fig 2). Rac1B is also an important player in matrix metalloproteinase-3 (MMP3)-induced EMT in breast, lung, and pancreas carcinomas3,6-8. Conversely, Rac1B can also exert anti-tumor effects in certain cancers (e.g., pancreatic ductal adenocarcinomas [PDAC]). In PDAC-derived cells, Rac1B inhibited EMT induced by TGF-β1, as determined by measuring changes in cell morphology, gene expression of EMT markers, signaling cascades downstream of TGF-β1 activation, cell migration, and growth inhibition following RNAi-induced exon 3b-targeted knockdown of Rac1B3,9,10. In addition, Rac1B offers potential as a therapeutic target as different GTPase inhibitor compounds are selective for it over Rac111. In addition, Rac1B may serve as a prognostic marker for some cancers (e.g., breast, colorectal, hepatocellular carcinoma, non-small cell lung cancer, pancreatic cancer, chronic pancreatitis, and thyroid cancer)12-14.
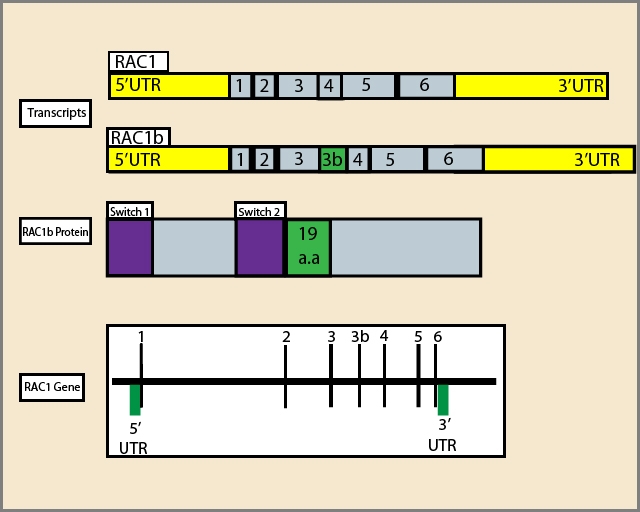 | 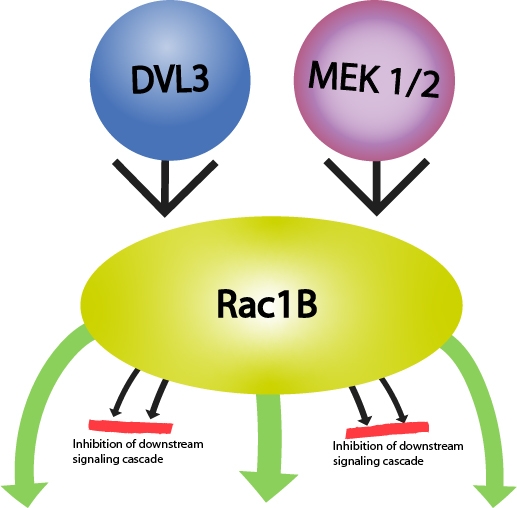 |
Figure 1: Diagram of RAC1 gene, alternative transcripts and RAC1b protein:1,2,3,4,5,6 respresent exons; a.a:amino acids. | Figure 2: Rac1B-mediated signaling relevant in tumorigenesis. Green arrows oriented outwards from Rac1B depict activation of a downstream signaling cascade, while those oriented towards Rac1B depict either activation (DVL3) or increased protein expression (MEK1/2) of a pathway. Red lines depict inhibition of the downstream signaling cascade. |
Upstream activators, downstream targets, and binding partners
Rac1B is an integral regulatory player in several signaling cascades implicated in cancer.(Fig 2) For example, Rac1B controls the activity of several kinases, including members of the mitogen-activated protein kinases which Rac1B negatively regulates3. Conversely, Rac1B activates NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells), a family of transcription factors that can enhance or inhibit tumorigenesis15, and it appears to do so in a manner that avoids activation of the RelB-mediated negative feedback pathway in colorectal cancer cells16. In thyroid cancer cells, Rac1B over-expression stimulates NFκB-mediated pro-proliferative and anti-apoptotic signaling pathways17. Additional signaling pathways impacted by Rac1B are Wnt/β-catenin and TGF-β signaling3. Interestingly, Rac1B does not interact with Rho-GDP dissociation inhibitor (Rho-GDI), while Rac1 does5,15. Rac1B can bind the GTPase binding domain of p21-activated kinase (PAK) in a GTP-dependent fashion, but not full length PAK15. In comparison with Rac1, Rac1B either does not bind or binds with a reduced affinity to Rho-family effectors such as Rho-GDI, GIT-1, and IQGAP. In contrast, it strongly binds those proteins [e.g., SmgGDS, RACK1, and p120(ctn)] involved in transcriptional regulation, cell-cell adhesion, and motility3,18. The inability of Rac1B to interact with Rho-GDI means that most of the Rac1B is bound to the plasma membrane in the absence of Rho-GDI-mediated sequestration. Thus, although cells express low levels of Rac1B, there is a greater proportion that is available for activation compared to Rac13,5,15. Furthermore, Rac1B is immune from ubiquitination at the plasma membrane (Rac1B cannot activate Jun-N-terminal kinase which mediates Rac1 ubiquitination)3,19.
Rac1B inhibits Rac1
Rac1B negatively regulates Rac1 activity20,21. In HeLa cells, Rac1B expression prevents PDGF-induced Rac1 activation, reduces the proportion of membrane-bound Rac1, and elevates Rho activity. As might be expected, these changes manifest as altered cell morphology and motility due to the altered actin cytoskeleton dynamics which are regulated by Rho and Rac GTPases and which underlie these basic cellular functions20. Similarly, siRNA-mediated knockdown of Rac1B in a pancreatic carcinoma cell line correlates with an increase in Rac1 protein levels10. Rac1 and Rac1B differentially modulate TGF-β1-induced cell migration with the former promoting TGF-β1-mediated increases in cell migration, while Rac1B inhibits it9,21.
Summary
Despite obvious advances in understanding the unique role of Rac1B in tumorigenesis, much remains undiscovered, and the full potential of Rac1B as a prognostic indicator and therapeutic target very much remains in question. Further differentiation of Rac1B versus Rac1 in cancer-associated signaling cascades is also of paramount importance. Several studies focused on elucidating Rac1B’s potential in these areas include development of a biosensor-based system that determines the ratio of Rac1/Rac1B in blood serum in real-time22 and studying the therapeutic feasibility of switching the ratio of Rac1/Rac1B in those cancers delineated by Rac1B over-expression3. To support this research, Cytoskeleton offers a wide range of GTPase reagents, including Rho-family GTPase activation assay kits, antibodies, GEF and GAP activity assay kits, live cell imaging probes for F-actin, and Signal Seeker kits for studying the post-translational modifications of target proteins up- and downstream of Rac1B.
References
1. Schnelzer A. et al. 2000. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, rac1b. Oncogene. 19, 3013-3020.
2. Fiegen D. 2004. Alternative splicing of rac1 generates rac1b, a self-activating gtpase. J. Biol. Chem. 279, 4743-4749.
3. Melzer C. et al. 2019. RAC1B: A Rho GTPase with versatile functions in malignant transformation and tumor progression. Cell. 8, 21.
4. Haeusler L.C. et al. 2006. Purification and biochemical properties of rac1, 2, 3 and the splice variant rac1b. Methods Enzymol. 406, 1-11.
5. Singh A. et al. 2004. Rac1b, a tumor associated constitutively active rac1 splice variant, promotes cellular transformation. Oncogene. 23, 9369–9380.
6. Lee K. et al. 2012. Matrix compliance regulates rac1b localization, nadph oxidase assembly, and epithelial-mesenchymal transition. Mol. Biol. Cell. 23, 4097–4108.
7. HChen Q.K. et al. 2013. Extracellular matrix proteins regulate epithelial–mesenchymal transition in mammary epithelial cells. Differentiation. 86, 126–132.
8. Mehner C. et al. 2014. Tumor cell-derived mmp3 orchestrates rac1b and tissue alterations that promote pancreatic adenocarcinoma. Mol. Cancer Res. 12, 1430–1439.
9. Ungefroren H. et al. 2014. Rac1b negatively regulates TGF-β1-induced cell motility in pancreatic ductal epithelial cells by suppressing Smad signalling. Oncotarget. 5, 277-290.
10. Witte D. et al. 2017. Negative regulation of TGF-β1-induced MKK6-p38 and MEK-ERK signalling and epithelial-mesenchymal transition by Rac1b. Sci. Rep. 7, 17313.
11. Beausoleil E. et al. 2009. Structure–activity relationship of isoform selective inhibitors of rac1/1b gtpaseucleotide binding. Bioorg. Med. Chem. Lett. 19, 5594–5598.
12. Alonso-Espinaco V. et al. 2014. Rac1b overexpression correlates with poor prognosis in kras/braf wt metastatic colorectal cancer patients treated with first-line folfox/xelox chemotherapy. Eur. J. Cancer. 50, 1973–1981.
13. Mehner C. et al. 2015. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer. 6, 480–489.
14. Guinney J. et al. 2015. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356.
15. Matos P. et al. 2003. Tumor-related alternatively spliced rac1b is not regulated by rho-gdp dissociation inhibitors and exhibits selective downstream signaling. J. Biol. Chem. 278, 50442-50448.
16. Matos P. and Jordan P. 2006. Rac1, but not rac1b, stimulates relb-mediated gene transcription in colorectal cancer cells. J. Biol. Chem. 281, 13724–13732.
17. Faria M. et al. 2017 Rac1b overexpression stimulates proliferation and nf-kb-mediated anti-apoptotic signaling in thyroid cancer cells. PLoS ONE. 12, e0172689.
18. Orlichenko L. et al. 2010. The 19-amino acid insertion in the tumor-associated splice isoform rac1b confers specific binding to p120 catenin. J. Biol. Chem. 285, 19153–19161.
19. Visvikis O. et al. 2008. Activated rac1, but not the tumorigenic variant rac1b, is ubiquitinated on Lys 147 through a jnk-regulated process. FEBS J. 275, 386–396.
20. Nimnual A.S. et al. 2010. Perturbation of cytoskeleton dynamics by the opposing effects of rac1 and rac1b. Small GTPases. 1, 89–97.
21. Melzer C. et al. 2017. The role of TGF-β and its crosstalk with RAC1/RAC1b signaling in breast and pancreas carcinoma. Cell Commun. Signal. 15, 19.
22. Sahu V. et al. 2016. Quantification of Rac1 and Rac1b in serum of non small cell lung cancer by label free real time assay. Clin. Chim. Acta. 460, 231–235
Related Products
G-LISA Kits
RhoA G-LISA Activation Assay (Luminescence format) (Cat. # BK121)
RhoA G-LISA Activation Assay Kit (Colorimetric format) (Cat. # BK124)
RhoA G-LISA Activation Assay Kit (Colorimetric format) (Cat. # BK124-S)
Rac1,2,3 G-LISA Activation Assay (Colorimetric format) (Cat. # BK125)
Rac1 G-LISA Activation Assay (Luminescence format) (Cat. # BK126)
Cdc42 G-LISA Activation Assay (Colorimetric format) (Cat. # BK127)
Cdc42 G-LISA Activation Assay (Colorimetric format) (Cat. # BK127-S)
Rac1 G-LISA Activation Assay Kit (Colorimetric Based) (Cat. # BK128)
Rac1 G-LISA Activation Assay Kit (Colorimetric Based) (Cat. # BK128-S)
RalA G-LISA Activation Assay Kit (Colorimetric Based) (Cat. # BK129)
Ras G-LISA Activation Assay Kit (Colorimetric Based) (Cat . # BK131)
Arf1 G-LISA Activation Assay Kit (Colorimetric Based) (Cat. # BK132)
Arf6 G-LISA Activation Assay Kit (Colorimetric Based) (Cat. # BK133)
RhoA / Rac1 / Cdc42 G-LISA Activation Assay Bundle 3 Kits (Cat. # BK135)
Total RhoA ELISA (Cat. # BK150)
Rho Family Small G-Protein Tools
Rac/Cdc42 Activator II (Cat # CN02)
Rho/Rac/Cdc42 Activator I (Cat # CN04)
Pull-Down Activation
Ras Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK008)
Ras Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK008-S)
RhoA / Rac1 / Cdc42 Activation Assay Combo Biochem Kit (bead pull-down format) (Cat. # BK030)
Arf1 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK032-S)
Arf6 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK033-S)
Cdc42 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK034)
Cdc42 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK034-S)
Rac1 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK035)
Rac1 Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK035-S)
RhoA Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK036)
RhoA Pull-down Activation Assay Biochem Kit (bead pull-down format) (Cat. # BK036-S)
RalA Activation Assay Biochem Kit (bead pull-down format) (Cat . # BK040)
GGA3-PBD Beads (Binds active Arf1 and Arf6 proteins) (Cat. # GGA07)
PAK-PBD beads (binds active Rac/Cdc42 proteins) (Cat. # PAK02)
Rhotekin-RBD beads (binds active Rho proteins) (Cat. # RT02)
Acti-Stain Phalloidins
Acti-stain 488 Phalloidin (Cat. # PHDG1)
Acti-stain 555 Phalloidin (Cat. # PHDH1)
Acti-stain 670 Phalloidin (Cat. # PHDN1)
Rhodamine Phalloidin (Cat. # PHDR1)
