SARS-CoV-2 infection alters the transcriptomic landscape of human airway cells
- By Cytoskeleton Inc. - Live Cell News
- Feb 16, 2021

To define the early viral and antiviral replication landscape within the respiratory tract Fiege et al. recently elucidated changes in normal human tracheal bronchial epithelial cells (nHTBE) in response to SARS-CoV-2 infection +/- remdesivir. Importantly, nHTBE cells endogenously express two key factors required for viral entry angiotensin-converting enzyme 2 (ACE2), and transmembrane serine protease 2 (TMPRSS2). After infection with a fluorescent SARS-CoV-2 clone, investigators used flow cytometry to identify cells based on clusters of differentiation markers and SiR-Tubulin (Cat. # CY-SC002). Several differentially-infected cell populations were identified with ciliated cells (SiR-Tubulin +, CD271-) being the most infected, secretory cells showing modest infection (CD66c+, SiR-Tubulin-, CD271-), and basal cells (CD271+, SiR-Tubulin-, CD66c-) being uninfected. Cell treatment with SARS-CoV-2-inhibiting remdesivir, which elicits delayed RNA chain termination during viral replication, significantly reduced detection of infected cells. Detection of viral RNA by single-cell RNA sequencing (scRNA-seq) recapitulated flow cytometry findings, and importantly, reveals that several cell types treated with remdesivir transcriptionally clustered with mock-infected cells indicating that: 1) gene expression is strongly altered by infection and 2) that remdesivir-induced reversion to uninfected transcription is not specific to cell type. Analysis of TMPRSS2 revealed that viral particle-rich cell types exhibited strong expression relative to basal cells. Inhibition of TMPRSS2 with camostat nearly abrogated infection in ciliated and secretory cells suggesting TMPRSS2-mediated entry is a major factor for infection. nHTBEs were assessed for type 1-3 interferon (IFN) induction and despite a relatively low number of cells expressing IFN, robust induction of interferon stimulated genes (ISGs) was observed in accord with a canonical antiviral state. Seq data also revealed that cells with the highest viral detection displayed relatively low levels of ISGs possibly revealing transcriptomic and antiviral immune dysfunction. Using cutting edge scRNA-seq and flow cytometry paired with the premiere microtubule dye SiR-Tubulin, this study was able to reveal several insights in SARS-CoV-2 infection including remdesivir efficiency, viral infection susceptibility by cell type, and that TMPRSS2 is a major factor in the viral infection process.
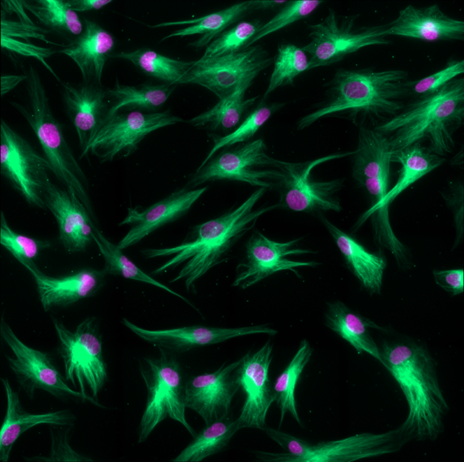
Caption 1: Live Human fibroblasts stained with SiR-tubulin (green) and Hoechst (magenta). ©Spirochrome, Switzerland
Link to Citation:
Related Products / Products Used: