K-Ras4B Mutant Protein: G12D+D38A (human recombinant with 6xHis-tag)
K-Ras4B Mutant Protein: G12D+D38A (human recombinant with 6xHis-tag)
* Limited stock available. If stock is not available, Cytoskeleton will produce a new batch upon request. Minimum order will apply. Inquire for more information.
Product Uses
- Study of G12D+D38A K-Ras4B exchange activity with different GEFs
- Identification of G12D+D38A K-Ras4B exchange factors (GEFs)
- Positive control for GEF studies
- Biochemical characterization of G12D+D38A K-Ras4B protein interactions
- Western blot standard
Materials
The G12D+D38A (glycine to aspartic acid at amino acid position 12) mutant human K-Ras4B protein has been produced in a bacterial expression system. The recombinant protein contains six histidine residues at its amino terminus (His-tag). The molecular weight of 6xHis tagged G12D+D38A K-Ras4B is approximately 25 kDa and it is supplied as a white lyophilized powder.
Storage
Before reconstitution, briefly centrifuge to collect the product at the bottom of the tube. The protein should be reconstituted to 5 mg/ml with the addition of 20 µl of ice cold nanopure water (100 µg size). When reconstituted, the protein will be in the following buffer: 50 mM Tris pH 7.5, 50 mM NaCl, 0.5 mM MgCl2, 5% (w/v) sucrose, and 1% (w/v) dextran. In order to maintain high biological activity of the protein, it is strongly recommended that the protein solution be supplemented with DTT to 1 mM final concentration, aliquoted into "experiment-sized" amounts, snap frozen in liquid nitrogen, and stored at -70°C. The protein is stable for six months if stored at -70°C. The protein should not be exposed to repeated freeze-thaw cycles. The lyophilized protein is stable at 4°C desiccated (<10% humidity) for one year.
Purity
Protein purity is determined by scanning densitometry of Coomassie Blue-stained protein on a 4-20% polyacrylamide gradient gel. His tagged G12D+D38A K-Ras4B protein was determined to be >85% pure. (see Figure 1).
Figure 1. G12D K-Ras4B Protein Purity Determination.

Legend: A 20 µg sample of recombinant G12D K-Ras4B protein (molecular weight approx. 25 kDa) was separated by electrophoresis in a 4-20% SDS-PAGE system and stained with Coomassie Blue. Protein quantitation was determined using the Precision Red Protein Assay Reagent (Cat. # ADV02). SeeBlue molecular weight markers are from Life Technologies Inc.
Biological Activity Assay
The biological activity of G12D K-Ras4B can be determined from the ability of the SOS1 exchange domain (SOS1-ExD) to catalyze the exchange of GDP for GTP onto mutant K-Ras4B. A standard biological assay for monitoring the biological activity of G12D K-Ras4B is an exchange assay utilizing the 2X Exchange Buffer from the RhoGEF exchange assay kit (Cat.# BK100) and the human SOS1 GEF domain (Cat.# CS-GE02). The Exchange Buffer contains Mant-GTP as reporter, which can be changed for Bodipy-GDP or -GTP for 490/513nm range and greater sensitivity.
Reagents
1. Recombinant G12D K-Ras4B protein (Cat.# CS-RS13)
2. Recombinant SOS1-ExD protein (Cat.# CS-GE02)
3. 2X Exchange Buffer (40 mM Tris pH 7.5, 100 mM NaCl, 20 mM MgCl2 , 0.1 mg/ml BSA, 1.5 µM mant-GTP)
4. Dilution Buffer (20 mM Tris pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.1 mg/ml BSA)
Equipment
1. Fluorescence spectrophotometer (λex=360 nm, λem=440 nm)
2. Corning 96-well half area plates (Cat. # 3686) or other plate with low protein binding surface.
Method
1. Dilute SOS1-ExD protein (Cat.# CS-GE02) to 1 μM (0.06 mg/ml) with Dilution Buffer.
2. Dilute G12D K-Ras4B to 40 µM (1.0 mg/ml) with Dilution Buffer.
3. Dissolve lyophilized 2X Exchange Buffer in 5 ml nanopure water and keep at room temperature.
4. Set up the plate reader for kinetic fluorescence measurements (Excitation wavelength at 360 nm and emission wavelength at 440 nm) with readings every 30 seconds for 30 minutes.
5. Add the following components together and mix well by gentle pipetting:
Association/Exchange reaction mix / 96 well black plate
2x Exchange Buffer / 50 μl
dH2O / 26 μl
40 µM G12D K-Ras4B/ 4 μl
6. Pipette 20 μl of 1 μM SOS1-ExD protein or Dilution Buffer into their respective wells and immediately pipette up and down twice and begin reading the fluorescence.
7. Once the readings are complete and the plate reader file has been saved, the exchange rate can be calculated by reducing the data to Vmax with the software that accompanies the plate reader.
Figure 2. GTP-exchange assay with KRas4B G12D and SOS1
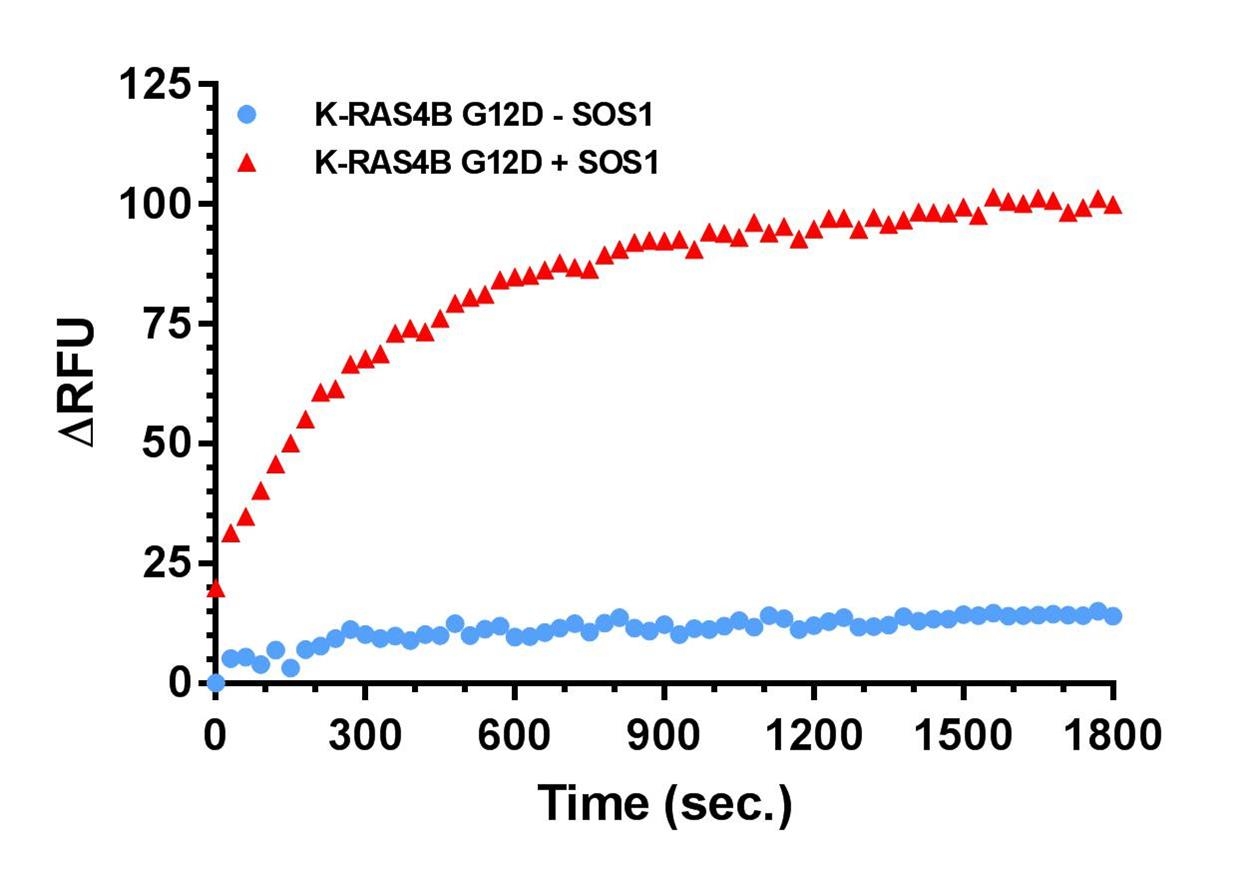
Legend: KRas4B G12D (Cat. # CS-RS13) was used in a nucleotide exchange reaction Bodipy-GTP from ThermoFisher was used as a reporter at 1 µM and KRas was at 2 µM. Ex/Em was 490/513nm. To initiate the exchange reaction, SOS1-ExD protein (red triangles), or Dilution Buffer (blue squares), was added to the wells, mixed, and fluorescence measurements were obtained using a Molecular Devices M2 SpectraMax. The average fluorescence data from quadruplicate assays were normalized to the fluorescence at the zero time point. The resulting data were plotted as the change in relative fluorescence units (ΔRFU) over time using GraphPad Prism software. ■ = K-Ras4B G12D no SOS1, and ▲ = K-Ras4B G12D with SOS1.
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com



