Dynein: Hard working, but team oriented
In eukaryote cells, cytoplasmic dynein complex (CDC) and kinesin-14 family members are the only two types of motors known to move cargo to the minus-end of microtubules (MTs)1. This function is critical for cell metabolism, structure, and movement. The majority of these processes utilize CDC because of its high fidelity and steady processive movement. CDC’s wide range of roles is impressive and the roles are further defined by ancillary proteins which select cargo or an intracellular location. Examples of the diversity of these processes are: a) transporting cargo into neuronal dendrites2, b) mitotic chromosome movement3, c) a pre-caspase pathway to apoptosis4, d) mitochondrial movement and fission5, and e) amyloid precursor protein vesicle transport6. Likewise, ancillary proteins for these functions are varied and include Presenilin, LIS1, NUDEL, NuMA, p150 Glued in dynactin, Miro, Milton, BimL, and BimEL, to name but a few.
Interestingly, the structure of CDC’s motor domain is much like a torsion gear design7 (Figure 1), which might represent its actual mechanism. Each dynein molecule is composed of a six domain ring motor head attached to a 20 nm amino terminal coiled-coil tail which binds the cargo and a 9 nm coiled-coil neck with a small MT binding domain at its end. The six domain ring binds up to four ATPs, with one domain hydrolyzing ATP in the power cycle and the other three poised to act as regulators or switches. Domains 5 and 6 do not contain the adenine binding motif (P-loop), which therefore does not allow ATP to bind. The torsion gear mechanism may occur due to the six domain ring being expanded at low loads and being compressed at high loads. At higher loads, the CDC may lie down on the MT surface and help create a catch-bond which stops movement in the plus-end direction.
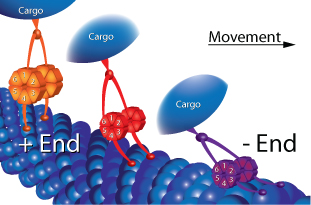
Figure 1: Schematic representation of proposed torsion gear mechanism and microtubule (MT) catch-bonding. The six member ring of CDC’s motor domain is shown in a relaxed state in orange and during high load conditions the ring is compressed as shown by the red model. During very high load conditions, CDC’s microtubule binding domain clamps down onto the MT as shown by the purple model.
There are several methods utilized by motors to move along MTs; these are single head one step, double head step over step, stutter step, and linear diffusion8-10. The most well-known movement is the step over step method utilized by double head motor proteins like kinesin heavy chain (KHC, kinesin-1 sub-family) and cytoplasmic myosin. These motors move without speed or gear control and more frequently fall off MTs compared to CDC under similar loads. In contrast, CDC is a different case; for example, in vitro, it moves in steps as large as 24-36 nm, which is equivalent to the length of 3 to 4 tubulin heterodimers11. This is surprising because CDC exists as a dimer with a spacer distance (motor to MT) of 9 nm each, but the motor domains are not bound closely to each other as they are in myosin and kinesin, and there is additional spacer length from the cargo binding spacer which is 15 nm long (Figure 1). In vitro, there is not much resistance placed on the motor in the form of cargo dragging through a cellular matrix. In vivo, the CDC movement is very different, with step length reduced to 8 nm when under load, which corresponds to the length of a tubulin heterodimer12. Thus, CDC can change its step size and maintain MT contact under different loads.
In recent years, some exquisite live cell microscopic work by several groups 11-15 has revealed how dynein moves and maintains control of movement in vivo. The groups improved on earlier work by a) utilizing laser-captured vesicles to measure force, b) by limiting their data to only straight tracking (<5% angles) particles, c) by more accurately defining what was a stall and a run in one direction, d) measuring the width of a stalled vesicle as an indication of opposing motors working against each other, and e) utilizing late endosome vesicles, which show more uniform motor protein components, i.e., KHC and CDC only.
When MT minus-end-directed vesicles are observed in vivo, they move in plus and minus-end directions and also stall; in effect the vesicles are changing from KHC to CDC driven motion and back again. This scenario is non-intuitive and we might expect only the presence of CDC. Mallik’s group studied this movement in more detail, initially utilizing a laser trap to calculate the force generated on a moving vesicle12. In the majority of cases, the authors measured a force of 5-6 piconewtons (pN) in the plus direction (KHC) and a 5-8 pN force in the minus direction (CDC). From in vitro measurements, it is known that these forces reflect the presence of one KHC dimer and 5 to 10 CDC complexes. The length of motion after starting a run was also measured, yielding a ratio of 5:1 (6 nm CDC/1.2 nm KHC), which is highly in favor of minus-end movement. Furthermore, the speed and step size after stall were measured and both were in favor of CDC in the minus-end direction by a factor of 1.5 to 2.0 fold. All of these aspects combined to produce overall motion in the minus-end direction. The authors summarized their findings by concluding that dynein is a tenacious team worker where each molecule contributes 1 pN in force12, extending others’ findings that dynein catch-bonds MTs and prohibits movement in the plus direction16.
In summary, dynein represents an evolutionarily unique mechanochemical-geared motor which applies strong forces by combining the effects of multiple motors. Dynein is tenacious and catch-bonds tightly to MTs to oppose KHC motor force. At Cytoskeleton, we are developing load and non-load MT stimulated CDC ATPase assays which can be used to identify inhibitors and enhancers such as that reported by Firestone17. Dynein protein is also available as a custom protein preparation (see New proteins... table), contact tservice@cytoskeleton.com for more information.
Related Research Tools and Services
| New proteins made to order... | Application | Module # | Amount | |||||
Dynein (cytoplasmic) | Microtubule stimulated ATPase assay | CS-DN01 | 1 x 50 µg | |||||
MKLP2 motor domain | Microtubule stimulated ATPase assay | 1 x 50 µg | ||||||
KIF7 motor domain | Microtubule stimulated ATPase assay | CS-KF51 | 1 x 100 µg | |||||
Myosin S1 fragment (cardiac) | Actin or thin filament stimulated ATPase assay | CS-MYS03 | 1 x 250 µg | |||||
Myosin S1 fragment (skeletal) | F-actin stimulated ATPase assay | CS-MYS04 | 1 x 250 µg | |||||
Myosin S1 fragment (smooth) | F-actin stimulated ATPase assay | CS-MYS05 | 1 x 250 µg | |||||
Myosin S1 fragment (non-muscle) | F-actin stimulated ATPase assay | CS-MYS06 | 1 x 250 µg | |||||
Actin Thin Filaments | Calcium sensitive F-actin stimulated myosin ATPase assay | CS-TFC01 | 1 x 1 mg | |||||
Tropomyosin / Tropomodulin | Actin thin filament preparation | CS-TT05 | 1 x 1 mg | |||||
| Kinesin and Dynein Proteins | Cat. # | Amount | ||||||
CENP-E Motor Domain Protein | CP01-A | 2 x 25 µg | ||||||
Chromokinesin Motor Domain Protein | CR01-A | 2 x 25 µg | ||||||
Eg5 Motor Domain Protein | EG01-A | 2 x 25 µg | ||||||
Eg5 Homolog BimC Motor Domain Protein | BM01-A | 2 x 25 µg | ||||||
Eg5 Homolog BimC Motor Domain Protein | EG02-A | 2 x 15 µg | ||||||
KIFC3 Motor Domain Protein | KC01-A | 2 x 25 µg | ||||||
KIF3C Motor Domain Protein | KF01-A | 2 x 25 µg | ||||||
Kinesin Heavy Chain Motor Domain Protein | KR01-A | 2 x 25 µg | ||||||
MCAK Motor Domain Protein | MK01-A | 2 x 25 µg | ||||||
MKLP1 Motor Domain Protein | MP01-A | 2 x 25 µg | ||||||
Kinesin ELIPA Biochem Kit | BK060 | 96 assays | ||||||
Kinesin ATPase Endpoint Assay | BK053 | 1000 assays | ||||||
Microtubules (pre-formed) | MT002 | 4 x 500 µg | ||||||
Actin Filaments (pre-formed) | AKF99 | 1 x 1 mg | ||||||
References
- Paschal B.M. & Vallee R.B. 1987. Isolated flagellar outer arm dynein translocates brain microtubules in vitro. Nature. 330, 672-674.
- Sasaki S. et al. 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 28, 681-696.
- Merdes A. et al. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87, 447-458.
- Puthalakath H. et al. 1999. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with dynein motor complex. Mol. Cell. 3, 287-296.
- Schwarz T.L. 2013. Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. doi 10.1101/cshperspect.a011304.
- Gunawardena S. et al. 2013. Presenilin controls kinesin-1 and dynein function during APP-vesicle transport in vivo. Hum. Mol. Genet. 22, 3828-2843.
- Vallee R.B. et al. 1988. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 332, 561-563.
- Hirokawa N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 279, 519-526.
- Kull F.J. & Endow S.A. 2013. Force generation by kinesin and myosin cytoskeletal motor proteins. J. Cell Sci. 126, 9-19.
- Vallee R.B. et al. 2012. Multiple modes of cytoplasmic dynein regulation. Nat. Cell Biol. 14, 224-230.
- DeWitt M.A. et al. 2012. Cytoplasmic dynein moves through uncoordinated stepping of AAA+ ring domains. Science. 335, 221-225.
- Rai A.K. et al. 2013. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 152, 172-182.
- Soppina V. et al. 2009. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 19381-19386.
- Qiu W. et al. 2012. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 19, 193-200.
- Dufrene Y.F. et al. 2011. Five challenges to bringing single-molecule force microscopy into live cells. Nat. Methods. 8, 123-127.
- Kunwar A. et al. 2011. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid droplet transport. Proc. Natl. Acad. Sci. U.S.A. 108, 18960-18965.
- Firestone A.J. 2012. Small molecule inhibitors of AAA+ ATPase motor cytoplasmic dynein. Nature. 484, 125-129.

