Laminin (Red fluorescent, rhodamine)
Product Uses Include
- Cell invasion assays (1)
- FACS analysis of laminin binding cells (2)
Material
Laminin-1 is purified from EHS tumor tissue and is free of the laminin binding protein entactin which is a common contaminant in some laminin preparations (150 kDa). Protein purity is determined by scanning densitometry of Coomassie Blue stained protein on a 4-20% polyacrylamide gel. The laminin is >90% pure (Figure 1).
The protein is modified to contain covalently linked rhodamines at random surface lysines. An activated ester of rhodamine [(5-(and 6)-carboxytetramethylrhodamine succinimidyl ester] is used to label the protein. Labeling stoichiometry is determined by spectroscopic measurement of protein and dye concentrations. Final labeling stoichiometry is 2-5 dyes per protein molecule (Figure 2). The material is guaranteed to contain <15% of free dye and >85% of dye conjugated to laminin. Rhodamine laminin can be detected using a filter set of 535 nm excitation and 585 nm emission.
Laminin runs as individual subunits on SDS-PAGE with an apparent molecular weight of 400 and 225 kDa (Figure 1). LMN01 is supplied as a pale pink lyophilized powder. Each vial of LMN01 contains 20 μg protein.
Purity
Purity is determined by scanning densitometry of proteins on SDS-PAGE gels. Samples are >90% pure.
Figure 1: Rhodamine Laminin Purity Determination
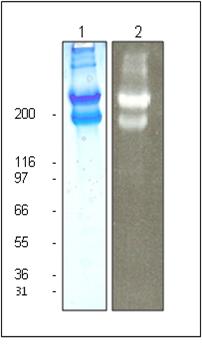
Legend: 20 μg of unlabeled laminin (Lane 1) and 20 μg of rhodamine laminin (Lane 2) was separated by electrophoresis in a 4-20% SDS-PAGE system. The unlabeled protein was stained with Coomassie Blue and visualized in white light. The rhodamine labeled protein was visual-ized under UV light. The alpha subunit runs at 400 kDa (top band) while the beta and gamma subunits run as a 225 kDa doublet (lower band). Protein quantitation was determined with the Precision Red™ Protein Assay Reagent (Cat. # ADV02). Mark12 molecular weight markers are from Invitrogen.
Figure 2: Detection of laminin (Red fluorescent, rhodamine)
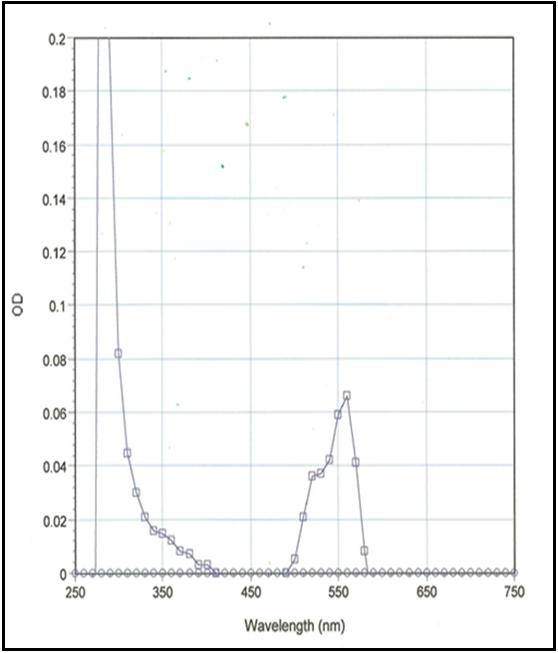
Legend: LMN01 was diluted with Milli-Q water and its absorbance spectrum was scanned between 250 and 750 nm. In this example, rhodamine labeling stoichiometry was calculated to be 2.7 dyes per laminin protein using the absorbancy maximum for rhodamine at 565 nm and the Beer-Lambert law. Dye extinction coefficient when protein bound is 70,000M-1cm-1 .
References
1. Kelly T. et al. 1994. Invadopodia promote proteolysis of a wide variety of extracellular matrix proteins. J. Cellular Physiol. 158: 299-308.
2. Tronchin G. et al. 1997. Expression and identification of a laminin-binding protein in Aspergillus fumigates conidia. Infection & Immunity 65: 9-15.
For product Datasheets and MSDSs please click on the PDF links below. For additional information, click on the FAQs tab above or contact our Technical Support department at tservice@cytoskeleton.com
Question 1: What is the optimal excitation and emission filter settings to visualize the rhodamine fluorescence?
Answer 1: Rhodamine-labeled laminin can be detected using a filter set of 535 nm excitation and 585 nm emission.
Question 2: What is the labeling stoichiometry?
Answer 2: Rhodamine labeling stoichiometry was calculated to be 2-5 dyes per laminin protein using the absorbancy maximum for rhodamine at 565 nm and the Beer-Lambert law. Dye extinction coefficient when protein bound is 70,000 M-1cm-1.
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com



