The Cytoskeleton’s Role In Autism Spectrum Disorder
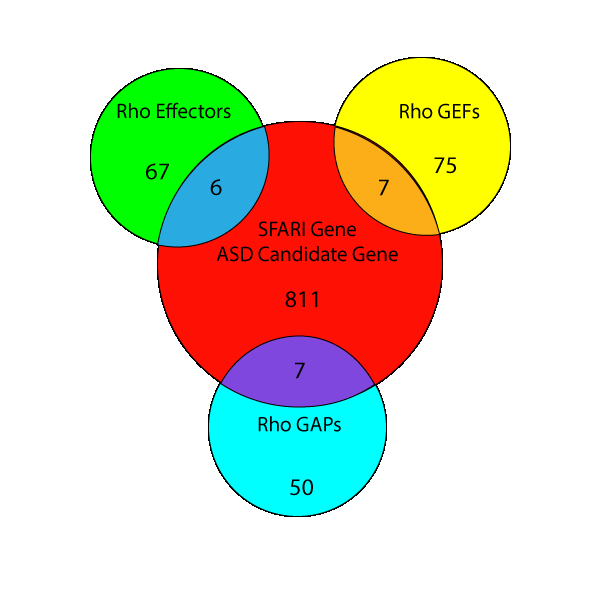
Above: Overlap of human gene sets of RhoGEFs, RhoGAPs, and Rho effectors with autism spectrum disorder (ASD) risk genes in Simons foundation autism research initiative (SFARI).
References:
1.Lord C, Brugha TS, Charman T, Cusack J, Dumas G, Frazier T, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6(1):5.
2.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180(3):568-84 e23.
3.Baron-Mendoza I, Maqueda-Martinez E, Martinez-Marcial M, De la Fuente-Granada M, Gomez-Chavarin M, Gonzalez-Arenas A. Changes in the Number and Morphology of Dendritic Spines in the Hippocampus and Prefrontal Cortex of the C58/J Mouse Model of Autism. Front Cell Neurosci. 2021;15:726501.
4.Guo D, Yang X, Shi L. Rho GTPase Regulators and Effectors in Autism Spectrum Disorders: Animal Models and Insights for Therapeutics. Cells. 2020;9(4).
5.Horn S, Au M, Basel-Salmon L, Bayrak-Toydemir P, Chapin A, Cohen L, et al. De novo variants in PAK1 lead to intellectual disability with macrocephaly and seizures.Brain.2019;142(11):3351-9.
6.Kernohan KD, McBride A, Hartley T, Rojas SK, Care4Rare Canada C, Dyment DA, et al. p21 protein-activated kinase 1 is associated with severe regressive autism, and epilepsy. Clin Genet. 2019;96(5):449-55.
7.Cheng C, Hou Y, Zhang Z, Wang Y, Lu L, Zhang L, et al. Disruption of the autism-related gene Pak1 causes stereocilia disorganization, hair cell loss, and deafness in mice. J Genet Genomics. 2021;48(4):324-32.
8.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209-15.
9.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216-21.
10.Nakashima M, Kato M, Matsukura M, Kira R, Ngu LH, Lichtenbelt KD, et al. De novo variants in CUL3 are associated with global developmental delays with or without infantile spasms. J Hum Genet. 2020;65(9):727-34.
11.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87(6):1215-33.
12.Morandell J, Schwarz LA, Basilico B, Tasciyan S, Dimchev G, Nicolas A, et al. Cul3 regulates cytoskeleton protein homeostasis and cell migration during a critical window of brain development. Nat Commun. 2021;12(1):3058.
13.Amar M, Pramod AB, Yu NK, Herrera VM, Qiu LR, Moran-Losada P, et al. Autism-linked Cullin3 germline haploinsufficiency impacts cytoskeletal dynamics and cortical neurogenesis through RhoA signaling. Mol Psychiatry. 2021;26(7):3586-613.