K-Ras: Oncogenic protein and anti-cancer drug target
The discovery of the K-Ras GTPase occurred during an in vitro assay screen with the active transformation of mouse NIH 3T3 fibroblasts. K-Ras is one of three Ras oncogenic isoforms (N- and H-Ras being the others). The human K-ras gene contains an alternative fourth coding exon, resulting in the production of two different splice variants termed K-Ras4A and K-Ras4B. The two isomorphic proteins differ by 25 amino acid residues at the carboxy terminus. Ras4A is considered a minor splice variant, meaning that the majority of oncogenic K-Ras research focuses on K-Ras4B. Within the K-Ras protein, there are four domains. The first domain includes 85 amino acids at the N-terminus which are found to be identical in H-, K-, and N-Ras. The following 80 amino acids form a second domain, showing less conservation (70-80%) between the Ras proteins. The third domain spans the rest of the K-Ras protein, except for the last four amino acids, and represents a hypervariable region. The highly conserved carboxy-terminal motif CAAX (where C stands for cysteine, A for any aliphatic residue, and X for any uncharged amino acid) is the result of post-translational modifications and forms the last domain. Depending on the splice variant, K-Ras consists of either 189 or 188 amino acids with a molecular weight of 21 kDa (for either variant).
Crystallography studies revealed that the catalytic domain spans residues 1 to 171, including the region involved in guanine nucleotide binding where residues 10-16 and 56-59 interact with b- and c-phosphate, and residues 116-119 and 152-165 interact with the guanine base. The so-called core effector region (located between residues 32-40), represents an essential element for all interactions with putative downstream effectors and the GTPase-activating proteins (GAPs). A region encompassing the last four amino acids at the C-terminus (residues 186-189) is essential for the attachment of K-Ras to the plasma membrane.
Like the other Ras isoforms, K-Ras proteins are localized to the inner plasma membrane, bind GDP and GTP and possess an intrinsic GTPase activity. K-Ras is involved in different types of ligand-mediated signal transduction pathways and influences proliferation, differentiation, transformation, and apoptosis by relaying mitogenic and growth signals into the cytoplasm and the nucleolus. K-Ras (like other Ras and Rho GTPases) cycles between an inactive (GDP-bound) and active (GTP-bound) state, acting as a molecular switch for many different signaling cascades and cellular processes. Activation is mediated guanine nucleotide exchange factors (GEFs) which stimulate the exchange of GDP for GTP. GTP is 10-fold more abundant in the cytosol than GDP. To turn off GTPase signaling, GAPs enhance the intrinsic GTPase activity of K-Ras. GAPs are necessary because the intrinsic GTPase activity is rather weak and not sufficiently effective for signal transduction pathways where rapid inactivation is required. In a normal cell, GAPs help to keep most of K-Ras in an inactive GDP-bound state.
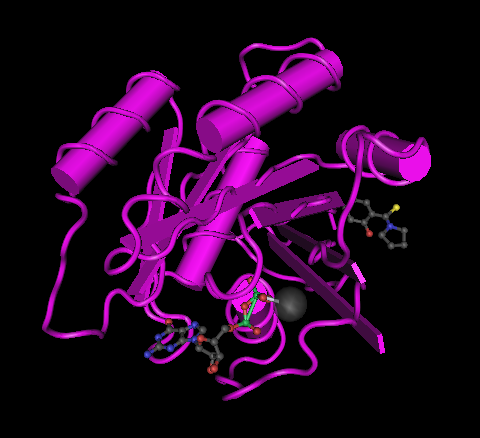
GDP-K-Ras complexed to a phenol moiety (small arrow head; PDB code 4EPT; 2-hydroxyphenyl)(pyrrolidin-1-yl)methanethione) as described in Sun Q. et al. 2012. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem. Int. Ed. Engl. 51, 6140–6143.
K-Ras in various human tumors harbors point mutations that confer transforming activity. Mutations leading to an amino acid substitution at the positions 12, 13, and 61 are the most common in naturally occurring (i.e., non-experimental) neoplasms and experimentally-induced animal tumors. K-Ras is considered the most relevant anti-cancer drug target as K-Ras mutations underlie 86% of Ras-linked cancers with 83% of K-Ras mutations at the G12 residue. For these reasons, K-Ras continues to be a primary target in anti-cancer drug studies. K-RAS mutations are found in a multitude of human cancers, with varying degrees of frequency, including pancreatic (80-90% frequency), colon and rectal (25-60%), lung (25-60%), prostate (0-25%), skin (0-25%), thyroid (0-60%), liver (10-25%), ovary (0-50%), endometrium (10-40%), kidney (0-50%), brain (0-15%), testis (10-45%), leukemia (5-15%), urinary/bladder (5%), head and neck (10%), and breast (10%).
For more information on K-Ras, please see these citations and references:
Papke B. and Der C.J. 2017. Drugging RAS: Know the enemy. Science. 355, 1158-1163.
Montalvo S.K. et al. 2017. Rationale for RAS mutation-tailored therapies. Future Oncol. 13, 263-271.
Stephen A.G. et al. 2014. Dragging ras back in the ring. Cancer Cell. 25, 272-281.
Related Products
SOS1 Protein (ExD Exchange Domain, aa564-1049, 6xHis tag) (Cat. # SOS1)
K-Ras4B Protein: wild-type (Cat. # RS03)
K-Ras4B Protein: G12V (Human recombinant) (Cat. # RS04)
Ras G-LISA Activation Assay Kit (Colorimetric Based) - 96 assays (Cat. # BK131)
Ras Pull-down Activation Assay Biochem Kit (bead pull-down format) - 50 Assays (Cat. # BK008)
