Controlling Ubiquitination is Key in the War with Bacteria
Introduction
Ubiquitination is a well-studied post-translational modification (PTM) that regulates a multitude of cellular events through its well-established role of targeting proteins for degradation via the proteasome. More recently, the ubiquitin (Ub) machinery has been shown to play a key role in targeting organelles such as mitochondria and endoplasmic reticulum for recycling in cooperation with the autophagic machinery. However, the Ub system does not only regulate the host-cell’s protein turnover; rather, increasing evidence shows that it plays a key role in the cells defense against invading bacterial pathogens (reviewed in [1]) . In nearly all cases, ubiquitination of these targets occur through the classical E1, E2, and E3 enzyme cascade to attach Ub to key lysine residues on target proteins and mark them for degradation. Here we describe the unique ways the Ub system is deployed to defend the cell against invading bacterial pathogens, and how these pathogens have fought back to dismantle the ubiquitin system in a battle of survival.
Ubiquitin system – a component of the host cellular defense program
Intracellular pathogens (e.g. S. Typhimurium, L. pneumophilia, S. Flexneri, etc.) that are capable of invading, residing, and replicating inside the host cell cause serious, human diseases. Host cells quickly recognize and destroy these invaders using its cell-autonomous innate immunity; a collection of processes that identify, incapacitate, and destroy these pathogens (reviewed in [1]). One very important process, antibacterial autophagy, is a selective, receptor mediated mechanism employed by the cell to restrict bacterial proliferation by encapsulating the pathogen and/or their associated vacuoles and delivering them to autolysosomes (reviewed in [2]). Antibacterial autophagy can be activated by multiple mechanisms including the Ub system, which is recognized by the p62, NDP52, and optinurin autophagy related receptors.
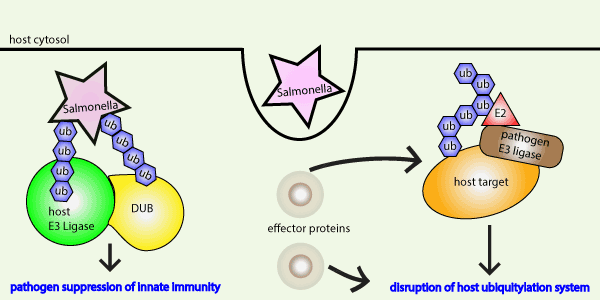
Fig 1 Legend: The Ub system is a critical component of the host cell innate immune response to bacterial invasion. Conversely, bacteria effector proteins hijack the ubiquitin machinery to suppress innate immunity.
The ability of Ub to form a coat on the surface of cytosolic bacteria S. Typhimurium and L monocytogenes was first discovered in the Brummel lab in 2004 [3]. Since then, investigators have focused on identifying the Ub machinery responsible for targeting invading bacteria, and with a particular interest towards the E3 ligase as these molecules play an essential role in target specificity. Several E3 ligases such as LUBAC, Parkin, LRSAM1 and others have now been shown to polyubiquitinate bacteria and target them for antibacterial autophagy (reviewed in [4]. Depletion of several of these ligases resulted in pronounced bacterial infection; for example, mice depleted of Parkin have an extreme susceptibility to M. tuberculosis infections [5]. Another ligase, Smurf1, was shown to be essential to limit bacterial load in mice and was shown to associate with bacteria in lungs of patients with pulmonary tuberculosis [6]. These studies revealed how E3 ligases function, their Ub chain preferences, and the bacteria they target. Still, it is clear there is much to learn about Ub-mediated antibacterial autophagy; such as, why there is a need for multiple ligases, the regulatory mechanisms that define the cooperative nature between these ligases, and what criteria dictates their recognition of specific bacteria.
Recently, Otten et al. found that the E3 ligase, RNF213, can specifically ubiquitinate lipopolysaccharides (LPS), a non-proteinaceous target, during Salmonella infection [7]. Importantly, this was one of the first specific bacterial substrates identified to be ubiquitinated. They showed that RNF213 is essential for the formation of the Ub coat through two distinct events; directly through ubiquitination of LPS and indirectly, through recruitment of a downstream E3 ligase, LUBAC, which forms M1-linked ubiquitin chains on bacteria [7]. Impairment of RNF213 resulted in bacteria that were not ubiquitinated, were not targeted by Ub-dependent autophagy receptors and did not undergo antibacterial autophagy. It will be noteworthy to identify the full spectrum of bacterial proteins and substrates that are targeted by the Ub system during Ub-dependent antibacterial autophagy. Relevant to this point, the bacterial cas9 protein has recently been identified as a target for SUMOylation and ubiquitination [8]. Cas9 is a well characterized bacterial protein that is a key member of the CRISPR machinery that is widely used in gene editing. Its native function in bacteria is as a defense mechanism against viruses. Thus, we wonder whether the Ub system recognizes Cas9 because it is a “foreign” protein target which should be degraded, or is there a specific mechanism that recognizes all bacterial proteins? Clearly, there is so much to learn and discover about this fascinating, innate immune mechanism.
Pathogens have evolved multiple mechanisms to deceive the ubiquitination process
There is a growing body of evidence that bacterial proteins undergo post-translational modifications at a higher than anticipated rate; more so, these modifications are important for many of the bacteria’s cellular processes (reviewed in [9, 10]). Select bacteria can even encode a prokaryotic ubiquitin like (PUP) protein that can target proteins to proteasomes, but these PUP proteins also perform other functions [11]. Multiple different bacteria have evolved mechanisms to circumvent and even hijack the host Ub system likely due to the selective pressure for survival. Bacterial pathogens such as S. Typhimurium, L pneumophilia, S. Flexneri, and enterohemorraghic E. coli have attained ubiquitin E3-like ligases that can transfer ubiquitin and manipulate the host Ub system (reviewed in [12]). For example, S. Typhimurium utilizes its HECT3-like ligase, SopA, to poly-ubiquitinate TRIM 56 and TRIM65 for K48 and target these proteins for degradation [13]. Ubiquitination is a reversible PTM that is kept in check by deubiquitinase (DUB) enzymes, and several bacteria have also evolved functional deubiquitinase-like proteins (reviewed in [12]). Interestingly these, OTU-like DUBs have chain preferences, and have been shown to cleave ubiquitin chains from bacterial containing vacuoles, leading to immune evasion. Recently, two new OUT-like DUBs were described in Legionella pneumophilia, and this structural study highlights key differences that may provide insight to their chain specificity [14]. Yan et al. recently identified another mechanism by which bacteria disrupts the Ub system to escape; specifically, Chromobacterium violaceum, utilizes its effector protein CteC to ADP-ribosylate ubiquitin itself on threonine 66 [15]. This PTM of Ub disrupted host cell polyubiquitination and was shown to be a key factor in its pathogenicity in mice. These are a few samples of the many ways bacteria have evolved to disrupt proper host cell ubiquitination to thwart host cell innate immune responses.
Summary
The host Ub system has been shown to play a critical role in innate immunity and is clearly a critical mechanism to protect cells against bacterial pathogens (see figure 1). This is not only evident by the number of ligases and other Ub machinery utilized by cells in Ub-dependent antibacterial autophagic mechanisms, but also based on the numerous bacterial like Ub ligases and DUBs that have evolved to circumvent or manipulate the host Ub system. It will be interesting to understand the interplay that occurs between bacterial effector proteins and the host Ub machinery as they compete for control of the Ub system, and how the host system responds as newly evolved Ub-escaping mechanisms from bacteria arise.
References
- Vozandychova, V., et al., The Ubiquitination System within Bacterial Host-Pathogen Interactions. Microorganisms, 2021. 9(3).
- Randow, F., How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy, 2011. 7(3): p. 304-9.
- Perrin, A.J., et al., Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol, 2004. 14(9): p. 806-11.
- Tripathi-Giesgen, I., C. Behrends, and A.F. Alpi, The ubiquitin ligation machinery in the defense against bacterial pathogens. EMBO Rep, 2021. 22(11): p. e52864.
- Manzanillo, P.S., et al., The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature, 2013. 501(7468): p. 512-6.
- Franco, L.H., et al., The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe, 2017. 22(3): p. 421-423.
- Otten, E.G., et al., Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature, 2021. 594(7861): p. 111-116.
- Ergunay, T., et al., Sumoylation of Cas9 at lysine 848 regulates protein stability and DNA binding. Life Sci Alliance, 2022. 5(4).
- Forrest, S. and M. Welch, Arming the troops: Post-translational modification of extracellular bacterial proteins. Sci Prog, 2020. 103(4): p. 36850420964317.
- Macek, B., et al., Protein post-translational modifications in bacteria. Nat Rev Microbiol, 2019. 17(11): p. 651-664.
- Pearce, M.J., et al., Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science, 2008. 322(5904): p. 1104-7.
- Franklin, T.G. and J.N. Pruneda, Bacteria make surgical strikes on host ubiquitin signaling. PLoS Pathog, 2021. 17(3): p. e1009341.
- Zhang, Y., et al., The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol, 2006. 62(3): p. 786-93.
- Shin, D., et al., Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection. Elife, 2020. 9.
- Yan, F., et al., Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination. Mol Cell, 2020. 78(4): p. 641-652 e9.
Signal Seeker™ PTM Detection Kits
New Signal-Seeker™ Acetyl-Lysine Detection Kit (30 assay) (Cat. # BK163)
New Signal-Seeker™ Acetyl-Lysine Detection Kit (10 assay) (Cat. # BK163-S)
Signal-Seeker™ Phosphotyrosine Detection Kit (30 assay) (Cat. # BK160)
Signal-Seeker™ Phosphotyrosine Detection Kit (10 assay) (Cat. # BK160-S)
Signal-Seeker™ Ubiquitination Detection Kit (30 assay) (Cat. # BK161)
Signal-Seeker™ Ubiquitination Detection Kit (10 assay) (Cat. # BK161-S)
Signal-Seeker™ SUMOylation 2/3 Detection Kit (30 assay) (Cat. # BK162)
Signal-Seeker™ SUMOylation 2/3 Detection Kit (10 assay) (Cat. # BK162-S)
Signal-Seeker™ SUMOylation 1 Detection Kit (30 Assay) (Cat. # BK165)
Signal-Seeker™ SUMOylation 1 Detection Kit (10 Assay) (Cat. # BK165-S)

