Acetyl Lysine Antibody Mouse Monoclonal
Anti-acetyl lysine antibody is a pan-acetyl lysine mouse monoclonal antibody that is part of the Signal-Seeker™ product line.The Anti-acetyl-lysine antibody recognizes proteins post-translationally modified by acetylation on the epsilon amine groups of lysine residues that occur on 30-50% of all proteins and in particular histones, p53, tubulin and myosin. A proprietary mixture of acetylated proteins was used to produce a highly robust antibody that has been shown to recognize a wide range of acetylated proteins in IP, WB, ChIP and IF applications. This Anti-acetyl-lysine antibody has many advantages when compared to other commercially available antibodies as shown below.
Validated Applications
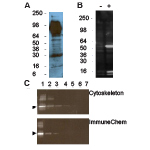
Western Blot using Acetyl Lysine Antibody
Cytoskeleton's Anti-Acetyl Lysine Antibody Cat. # AAC01 recognizes 0.005 ng of chemically acetylated BSA and is comparable in sensitivity to other commercially available antibodies. Unlike other Anti-Acetyle Lysine antibodies this antibody does not show cross reactivity with non-acetylated BSA. To see the full Western blot comparison, see the Optimized Protocols or the product datasheet.
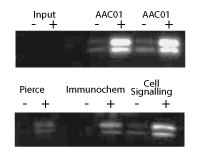
Immunoprecipitation using Acetyl Lysine Antibody
Ability of AAC01 to IP histones was compared to other commercially available antibodies. Cytoskeleton's Anti-Acetyl Lysine Antibody provides clear advantages for IP applications. Each tube is sufficient for approximately 20 IP assays. To see the full Immunoprecipitation comparison, see the Optimized Protocols or the product datasheet.
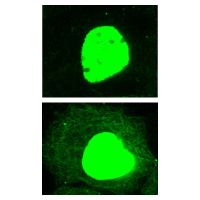
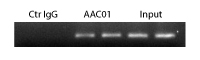
Chromatin was prepared from A431 cells, either untreated or TSA-treated (5 mM, 4 hrs). Briefly, cells were fixed with 1% formaldehyde for 10 min and enzymatically-sheared chromatins were immunoprecipitated by using anti-acetyl antibodies (1:100 dilution). The promoter region of housekeeping gene GAPDH was amplified by a primer pair and PCR products were analyzed by 2% agarose gel-electrophoresis. To see the recommended ChIP protocol, see the Optimized Protocols or the product datasheet.
Protein Acetylation Background
Acetylation of proteins can occur as a co-translational or post-translational modification (PTM) (1). Co-translational acetylation occurs at the N-terminal of approximately 85% of mammalian proteins, it is irreversible and is thought to be important in protein stability, localization and synthesis (1). Post-translational acetylation occurs on the epsilon amino group of lysine residues as a reversible and highly dynamic PTM that is known to be a key regulator in multiple cellular events, including chromatin structure, transcription, metabolism, signal transduction and cytoskeletal regulation (2-3). To date over 4,000 proteins have been identified as targets for PTM acetylation which is comparable to phosphorylation in cellular prevelance (3). Antibody AAC01 detects acetyl lysine PTMs.
References
1 Bogdan P. and Sherman F. 2002. The diversity of acetylated proteins. Genome Biol. 3 (5): reviews 0006.
2 Lundby A. et al. 2012. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and cellular patterns. Cell Reports 2:419-431.
3 Sadoul K. et al. 2010. The tale of protein lysine acetylation in the cytoplasm. J. Biomed. Biotech. 2011:1-15.
4 Golemis EA et. Al, Protein-Protein Interactions, CSHLP, 2005, p67
For more information contact: signalseeker@cytoskeleton.com
Associated Products:
Signal-Seeker™: BlastR™ Rapid Lysate Prep Kit (Cat. # BLR01)
For product Datasheets, MSDSs, and COAs please click on the PDF links below.
Sample Size Datasheet (Cat. AAC01-S): ![]()
Certificate of Analysis: Lot 013
Visit our Signal-Seeker™ Tech Tips and FAQs page for technical tips and frequently asked questions regarding this and other Signal-Seeker™ products click here
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com