Visualization of the First Tubulin Structures
The first images of tubulin within the cell began to be observed in the 1950s and 1960s via transmission electron microscopy(1). However, initial fixation methods at that time did not preserve the tubulin structure for it to be observed consistently. Initially, scientists thought these tubulin structures were canaliculi, endoplasmic reticulum, or filamentous elements(2,3). In 1963, after the development of using glutaraldehyde as a fixative, the consistency and resolution of tubulin improved, and the term “microtubules” (MTs) was introduced(4-6). Several years later, Gary Borisy and Ed Taylor at the University of Chicago used tritium-labeled colchicine as a small molecule probe to elucidate how colchicine inhibited mitosis. They discovered that a structural protein must be binding the labeled colchicine, as it was taken up by KB and Hela cells in tissue culture(7), sea urchin eggs(8), sea urchin sperm tails(9), and the brain(10). Soon after, Hideo Mohri gave the colchicine-binding protein the official name of tubulin(11). Interest in microtubules increased, as these cylindrical polymer structures were recognized as essential components of the cytoskeleton required for cell growth and maintaining cell shape(12).
In the early 1970’s it was revealed that tubulin was composed of a heterodimer of two closely related 55 kDa proteins called a- and b-tubulin(13-15). These two proteins are encoded by separate genes, or small gene families, whose sequences are highly conserved throughout the eukaryotic kingdom(16). Tubulin is a polar structure that can polymerize from both ends in vitro, however; the rate of polymerization is not equal. It has therefore become the convention to call the rapidly polymerizing end the plus (+) end and the slowly polymerizing end the minus (-) end. When tubulin polymerizes it initially forms protofilaments, MTs consist typically of 13 protofilaments that are 25 nm in diameter. Each µm of MT length is composed of 1650 heterodimers as observed with electron micrographs(17). Tim Mitchison and Marc Kirschner in 1984 found that unlike other polymers like F-actin in the cytoskeleton, microtubules can grow and suddenly shrink when they lose their GTP-caps through GTP hydrolysis(18,19). This GTP hydrolysis leads to the dynamic behavior of growth and shrinkage of tubulin known as dynamic instability. Recent advances in structural biology have now allowed research groups to examine MT dynamics at the atomic level.
The First High-Resolution Structure of Tubulin
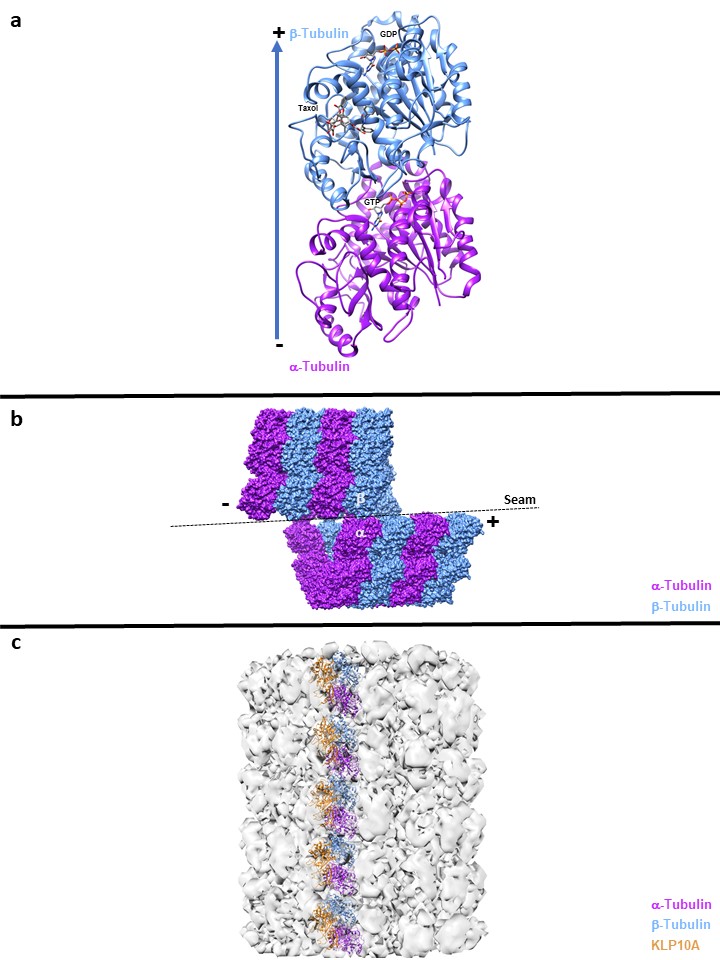
X-ray crystallography has been the gold standard for determining the three-dimensional molecular structure of proteins. High-resolution structures can be obtained at resolutions in the 1 to 2.5 Å (0.1 to 0.25 nm) range with X-ray crystallography. For tubulin, the first atomic high-resolution 2D crystalline structure of the ab-tubulin dimer (Fig 1a.) was determined by Nogales at 3.7 Å with electron microscopy using tubulin isolated from bovine brain produced in their lab and obtained from Cytoskeleton(20). The crystalline structure was obtained by using zinc-induced tubulin sheets stabilized by Taxotere, a chemical analog to Taxol. This structure revealed that each subunit is bound to a nucleotide, where the b-subunit contains the exchangeable and exposed GTP/GDP site (E-site), while the non-exchangeable GTP site (N-site) is located on the a-subunit and is buried within the dimer. This structure was also vital in revealing how Taxol stabilized MTs, by binding within a specific hydrophobic site on b-tubulin, with further structural analysis showing that Taxol stabilizes protofilament interactions(21). This structure provided a structural understanding of MT nucleotide exchange and insight into dynamic instability.
Figure legend:
A: High Resolution cyrstallography of αß tubulin dimer
B: α-α and ß-ß lateral interaction, except at the seam where the lateral contacts are α-ß and ß-α
C: Microtubule destabilization, the Microtubule depolymerase Kinesin-13s KLP10A bound to Microtubule structures induces tubulin curvature when the kinesin motor domain is bound to ATP
Improvements in Cryo-EM for Atomic-Resolution Structures of Microtubules
Obtaining atomic-resolution structures of MT polymers is necessary to further elucidate MT dynamics, but elucidating these structures with X-ray crystallography is not amenable. The filamentous nature of MTs makes them an ideal candidate to be studied by cryo-electron microscopy (Cryo-EM). However, initial resolutions of Cryo-EM MT structures were limited to resolutions of 20-25 Å(22 (2 and 2.5 nm)(22), where very little details of the MTs, such as the seam, could not be observed. Resolutions of MTs improved to 8-10 Å(21), but advancements in detector technology, image processing, and techniques have allowed Cryo-EM to undergo a “resolution revolution” where near-atomic resolutions of MTs down to approximately 3.5 Å can be obtained(23). These higher resolution structures have provided snapshots of MT dynamics in a solution-like state, unlike the zinc-induced anti-parallel tubulin sheets of the X-ray crystal structure.
The improved resolution of Cryo-EM revealed how protofilaments are arranged in MTs. The lateral interactions between protofilaments involve a-a and b-b contacts, except at the seam, where lateral contacts are a-b and b-a (Fig 1b)(24). These lateral interactions are important, as they are integral for determining the curvature of MTs. Calculating the curvature of MT structures has been of interest in the field, as it was believed that “straight” GTP-bound tubulin promotes stability and polymerization, while “curved” GDP-bound tubulin creates instability and destabilization of the MT lattice. However, this theory likely involves additional conformations, as the Cryo-EM structure of MTs in a possible conformational intermediate state bound to GMPCPP (GTP-like), showed a radial bend of approximately 5o compared to the 12o bend of the GDP-bound structure (25). A more recent structure also showed that GTP-hydrolysis initiates a conformational change that results in compression and strain to the MT lattice(26). Further work is needed to better understand the conformational changes that occur for a given nucleotide state and how these atomic models can provide additional answers for MT dynamics.
Microtubule-Associated Proteins and Microtubule Structures
The discovery of microtubule-associated proteins (MAPs) in the mid-1970s (28-31) revealed proteins factors that are capable of regulating MT dynamics. The improvements in Cryo-EM have started to reveal structures of MAPs bound to MTs. Understanding how MAPs can selectively discriminate different conformations of MTs to regulate MT dynamics is of great interest to the field. The Tau family proteins promote MT stabilization, where detailed structures of MT-Tau and MAP4-Kinesin1-MT complexes were solved(32,33). These structures show the advantages of using Cryo-EM, as the tau family MAPs are usually not captured, as they are disordered in solution and form aggregates. For MT destabilization, the MT depolymerase Kinesin-13s KLP10A (Fig 1c) bound to MTs structures was found to induce tubulin curvature when the kinesin motor domain is bound to ATP(34,35) and helps illustrate the complexity of mechanisms required to regulate MT dynamics.
Conclusion
These studies highlight the advancements from the first visual images of MTs from the 1950s to the recent atomic-resolution structures of MTs and their binding partners. Future studies will elucidate additional mechanisms involved with MT dynamics. Tubulin purified at Cytoskeleton Inc. has been used by several research groups cited in this newsletter. In addition to tubulin, Cytoskeleton Inc. offers other reagents to aid investigators in their research to help expand our knowledge in the tubulin field.
Citations
- Masters, Barry R (March 2009) History of the Electron Microscope in Cell Biology. In: Encyclopedia of Life Sciences (ELS). John Wiley & Sons, Ltd: Chichester.
- Wells WA. Microtubules Get a Name. J Cell Biol.2005;168(6):852–853.
- Borisy G, Heald R, Howard J, Janke C, Musacchio A, Nogales E. Microtubules: 50 years on from the discovery of tubulin. Nat Rev Mol Cell Biol. 2016;17(5):322-328.
- Slautterback DB. Cytoplasmic Microtubules. I. Hydra. J Cell Biol. 1963;18(2):367-388.
- Ledbetter MC, Porter KR. A "Microtubule" In Plant Cell Fine Structure. J Cell Biol. 1963;19(1):239-250.
- Sabatini DD, BenschK, Barrnett RJ. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963;17(1):19-58.
- Borisy GG, Taylor EW. The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J Cell Biol. 1967;34(2):525-33.
- Borisy GG, Taylor EW. The mechanism of action of colchicine. Colchicine binding to sea urchin eggs and the mitotic apparatus. J Cell Biol. 1967;34(2):535-48.
- Shelanski ML, Taylor EW. Isolation of a protein subunit from microtubules. J Cell Biol. 1967;34(2):549-554.
- Weisenberg RC, Borisy GG, Taylor EW. The colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry. 1968;7(12):4466-79.
- Mohri H. Amino-acid Composition of “Tubulin” constituting Microtubules of Sperm Flagella. Nature 1968;217:1053–1054.
- Tilney, Lewis G. and Keith R. Porter. “Studies On The Microtubules In Helioza: II. The Effect of Low Temperature on These Structures in the Formation and Maintenance of the Axopodia.” Journal of Cell Biology1967;34:327-343.
- Stephens RE. Thermal fractionation of outer fiber doublet microtubules into A- and B-subfiber components: A- and B-tubulin. J Mol Bio 1970;47(3):353-363.
- Jacobs M, McVittie A. Identification of the flagellar proteins of Chlamydomonas reinhardii, Experimental Cell Research 1970;70(1):53-61.
- Heterogeneity of Tubulin Subunits Feit H, Slusarek L, Shelanski Heterogeneity of Tubulin Subunits PNAS 1971;68(9):2028-2031.
- Little M, Krauhs E, Ponstingl H. Tubulin sequence conservation. Biosystems. 1981;14(3-4):239-46.
- Amos LA, Klug A. Arrangement of Subunits in Flagellar Microtubules. J Cell Sci1974;14(3):523–549.
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature 1984;312:232–237.
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature 1984;312:237–242.
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 1998;391(6663):199-203.
- Li, H, DeRosier DJ, Nicholson WV, Nogales, E. Downing, KH. Microtubule structure at 8 A resolution. Structure 2002;10:1317–1328.
- Nogales E, Kellogg EH. Challenges and opportunities in the high-resolution cryo-EM visualization of microtubules and their binding partners. Curr Opin Struct Biol. 2017;46:65-70.
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 2015;162:849–859.
- Nogales E, Zhang R. Visualizing microtubule structural transitions and interactions with associated proteins. Curr Opin Struct Biol. 2016;37:90-6.
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature 2005;435(7044):911-915.
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 2014;157(5):1117-29.
- Brouhard GJ, Rice LM. The contribution of αβ-tubulin curvature to microtubule dynamics. J Cell Biol. 2014;207(3):323-34.
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. PNAS 1975;72(5):1858-62.
- Sloboda RD, Dentler WL, Rosenbaum JL. Microtubule-associated proteins and the stimulation of tubulin assembly in vitro. Biochemistry 1976;15(20):4497-505.
- Murphy DB, Borisy GG Association of high-molecular-weight proteins with microtubules and their role in microtubule assembly in vitro. PNAS 1975;72(7):2696-700.
- Sloboda RD, Rudolph SA, Rosenbaum JL, Greengard P. Cyclic AMP-dependent endogenous phosphorylation of a microtubule-associated protein. PNAS 1975;72(1):177-81.
- Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E. Near-atomic model of microtubule-tau interactions. Science 2018;360(6394):1242-1246.
- Shigematsu H, Imasaki T, Doki C, Sumi T, Aoki M, Uchikubo-Kamo T, Sakamoto A, Tokuraku K, Shirouzu M, Nitta R. Structural insight into microtubule stabilization and kinesin inhibition by Tau family MAPs. J Cell Biol. 2018;217(12):4155-4163.
- Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 2013;3(3):759-68.
- Benoit, M.P.M.H., A.B. Asenjo, and H. Sosa. Cryo-EM Reveals the Structural Basis of Microtubule Depolymerization by Kinesin-13s. Nature Comm. 2018;9:1-13.
Related Products:
Pre-formed Microtubules
Purified Tubulins and MAPS
Tubulin protein (97% pure): porcine brain (Cat. # HTS03)
Tubulin Kits
Tubulin polymerization HTS assay using >97% pure tubulin, OD based - Porcine (Cat. # BK004P)
Tubulin polymerization assay using >99% pure tubulin, OD based - Porcine (Cat. # BK006P)
Tubulin polymerization assay using >99% pure tubulin, fluorescence based (Cat. # BK011P)
Microtubule Binding Protein Spin-Down Assay Biochem Kit (Cat. # BK029)
Microtubule/Tubulin In Vivo Assay Biochem Kit (Cat. # BK038)