| Author | Title | Journal | Year | Article Link |
|---|
| Nithianantham, Stanley et al. | The kinesin-5 tail and bipolar minifilament domains are the origin of its microtubule crosslinking and sliding activity | Molecular biology of the cell | 2023 | ISSN 1939-4586 |
| McHugh, Toni et al. | Potent microtubule-depolymerizing activity of a mitotic Kif18b–MCAK–EB network | Journal of Cell Science | 2023 | ISSN 1477-9137 |
| Yang, Shuzhen et al. | EB1 decoration of microtubule lattice facilitates spindle-kinetochore lateral attachment in Plasmodium male gametogenesis | Nature Communications 2023 14:1 | 2023 | ISSN 2041--1723 |
| Hoshino, Asumi et al. | The microtubule-severing protein UNC-45A preferentially binds to curved microtubules and counteracts the microtubule-straightening effects of Taxol | Journal of Biological Chemistry | 2023 | ISSN 1083-351X |
| Oda, Haruka et al. | Actin filaments accumulated in the nucleus remain in the vicinity of condensing chromosomes in the zebrafish early embryo | Biology Open | 2023 | Article Link |
| Palumbo, Jacob et al. | Directly Measuring Forces Within Reconstituted Active Microtubule Bundles | JoVE (Journal of Visualized Experiments) | 2022 | ISSN 1940--087X |
| Qian, Pengge et al. | Apical anchorage and stabilization of subpellicular microtubules by apical polar ring ensures Plasmodium ookinete infection in mosquito | Nature Communications 2022 13:1 | 2022 | ISSN 2041--1723 |
| Henty-Ridilla, Jessica L. | Visualizing Actin and Microtubule Coupling Dynamics In Vitro by Total Internal Reflection Fluorescence (TIRF) Microscopy | JoVE (Journal of Visualized Experiments) | 2022 | ISSN 1940--087X |
| Sasanpour, Mehrzad et al. | Reconstituting and Characterizing Actin-Microtubule Composites with Tunable Motor-Driven Dynamics and Mechanics | JoVE (Journal of Visualized Experiments) | 2022 | ISSN 1940--087X |
| Planelles-Herrero, Vicente Jose et al. | Elongator stabilizes microtubules to control central spindle asymmetry and polarized trafficking of cell fate determinants | Nature Cell Biology | 2022 | ISSN 1476--4679 |
| Kuzmić, Mira et al. | Septin-microtubule association via a motif unique to isoform 1 of septin 9 tunes stress fibers | Journal of Cell Science | 2022 | ISSN 1477-9137 |
| Lee, Gloria et al. | Myosin-driven actin-microtubule networks exhibit self-organized contractile dynamics | Science Advances | 2021 | ISSN 2375-2548 |
| Hough, Cameron M. et al. | Disassembly of microtubules by intense terahertz pulses | Biomedical Optics Express | 2021 | ISSN 2156--7085 |
| Habicht, Juri et al. | UNC-45A breaks the microtubule lattice independently of its effects on non-muscle myosin II | Journal of Cell Science | 2021 | ISSN 1477-9137 |
| Kundu, Tanushree et al. | Coupling of dynamic microtubules to F-actin by Fmn2 regulates chemotaxis of neuronal growth cones | Journal of Cell Science | 2021 | ISSN 1477-9137 |
| Saper, Gadiel et al. | Robotic end-to-end fusion of microtubules powered by kinesin | Science Robotics | 2021 | ISSN 2470-9476 |
| Alfieri, Angus et al. | Two modes of PRC1-mediated mechanical resistance to kinesin-driven microtubule network disruption | Current Biology | 2021 | ISSN 1879-0445 |
| Kaur, Simranpreet et al. | Expansion of the phenotypic spectrum of de novo missense variants in kinesin family member 1A (KIF1A) | Human Mutation | 2020 | ISSN 1098-1004 |
| Ricketts, Shea N. et al. | Triggering Cation-Induced Contraction of Cytoskeleton Networks via Microfluidics | Frontiers in Physics | 2020 | ISSN 2296-424X |
| Pyrpassopoulos, Serapion et al. | Modulation of Kinesin’s Load-Bearing Capacity by Force Geometry and the Microtubule Track | Biophysical Journal | 2020 | ISSN 0006--3495 |
| Aher, Amol et al. | CLASP Mediates Microtubule Repair by Restricting Lattice Damage and Regulating Tubulin Incorporation | Current Biology | 2020 | ISSN 1879-0445 |
| Chen, Keyu et al. | Giant ankyrin-B suppresses stochastic collateral axon branching through direct interaction with microtubules | Journal of Cell Biology | 2020 | ISSN 1540-8140 |
| Rodríguez-García, Ruddi et al. | Mechanisms of Motor-Independent Membrane Remodeling Driven by Dynamic Microtubules | Current Biology | 2020 | ISSN 1879-0445 |
| Saper, Gadiel et al. | Kinesin-propelled label-free microtubules imaged with interference reflection microscopy | New Journal of Physics | 2020 | ISSN 1367-2630 |
| Adriaans, Ingrid E. et al. | MKLP2 Is a Motile Kinesin that Transports the Chromosomal Passenger Complex during Anaphase | Current Biology | 2020 | ISSN 1879-0445 |
| Gaska, Ignas et al. | The Mitotic Crosslinking Protein PRC1 Acts Like a Mechanical Dashpot to Resist Microtubule Sliding | Developmental Cell | 2020 | ISSN 1878-1551 |
| Kalra, Aarat P. et al. | Investigation of the electrical properties of microtubule ensembles under cell-like conditions | Nanomaterials | 2020 | ISSN 2079-4991 |
| Tuszynski, Jack A. et al. | Microtubules as Sub-Cellular Memristors | Scientific Reports | 2020 | ISSN 2045-2322 |
| Francis, Madison L. et al. | Non-monotonic dependence of stiffness on actin crosslinking in cytoskeleton composites | Soft Matter | 2019 | ISSN 1744-6848 |
| Leong, Su Ling et al. | Reconstitution of Microtubule Nucleation In Vitro Reveals Novel Roles for Mzt1 | Current Biology | 2019 | ISSN 0960-9822 |
| Lopes, Joseph et al. | Membrane mediated motor kinetics in microtubule gliding assays | Scientific Reports | 2019 | ISSN 2045-2322 |
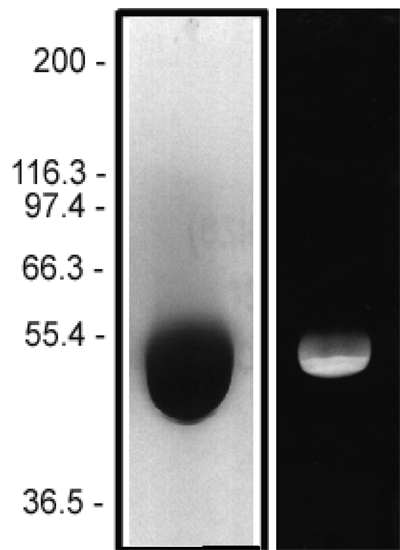
| Faltova, Lenka et al. | Crystal Structure of a Heterotetrameric Katanin p60:p80 Complex | Structure | 2019 | ISSN 1878-4186 |
| Grueb, Saskia S. et al. | The formin Drosophila homologue of Diaphanous2 (Diaph2) controls microtubule dynamics in colorectal cancer cells independent of its FH2-domain | Scientific Reports | 2019 | ISSN 2045-2322 |
| Chudinova, Elena M. et al. | On the interaction of ribosomal protein RPL22e with microtubules | Cell Biology International | 2019 | ISSN 1095-8355 |
| Fu, Meng meng et al. | The Golgi Outpost Protein TPPP Nucleates Microtubules and Is Critical for Myelination | Cell | 2019 | ISSN 1097-4172 |
| Ricketts, Shea N. et al. | Varying crosslinking motifs drive the mesoscale mechanics of actin-microtubule composites | Scientific Reports | 2019 | ISSN 2045-2322 |
| Nakos, Konstantinos et al. | Septin 2/6/7 complexes tune microtubule plus-end growth and EB1 binding in a concentration- And filament-dependent manner | Molecular Biology of the Cell | 2019 | ISSN 1939-4586 |
| Karasmanis, Eva P. et al. | Erratum: Polarity of Neuronal Membrane Traffic Requires Sorting of Kinesin Motor Cargo during Entry into Dendrites by a Microtubule-Associated Septin (Developmental Cell (2018) 46(2) (204–218.e7), (S1534580718304982) (10.1016/j.devcel.2018.06.013)) | Developmental Cell | 2018 | ISSN 1878-1551 |
| Ganguly, Anindya et al. | Importin-β Directly Regulates the Motor Activity and Turnover of a Kinesin-4 | Developmental Cell | 2018 | ISSN 1878-1551 |
| Fan, Yuanwei et al. | The Arabidopsis SPIRAL2 Protein Targets and Stabilizes Microtubule Minus Ends | Current Biology | 2018 | ISSN 0960-9822 |
| Romé, Pierre et al. | A novel microtubule nucleation pathway for meiotic spindle assembly in oocytes | Journal of Cell Biology | 2018 | ISSN 1540-8140 |
| Colin, Alexandra et al. | Actin-Network Architecture Regulates Microtubule Dynamics | Current Biology | 2018 | ISSN 0960-9822 |
| Rao, Lu et al. | Combining structure–function and single-molecule studies on cytoplasmic dynein | Methods in Molecular Biology | 2018 | ISSN 1064-3745 |
| Aher, Amol et al. | CLASP Suppresses Microtubule Catastrophes through a Single TOG Domain | Developmental Cell | 2018 | ISSN 1878-1551 |
| Hilton, Nicholas A. et al. | Identification of TOEFAZ1-interacting proteins reveals key regulators of Trypanosoma brucei cytokinesis | Molecular Microbiology | 2018 | ISSN 1365-2958 |
| Höing, Susanne et al. | Dynarrestin, a Novel Inhibitor of Cytoplasmic Dynein | Cell Chemical Biology | 2018 | ISSN 2451-9448 |
| Reinemann, Dana N. et al. | Processive Kinesin-14 HSET Exhibits Directional Flexibility Depending on Motor Traffic | Current Biology | 2018 | ISSN 0960-9822 |
| Ricketts, Shea N. et al. | Co-Entangled Actin-Microtubule Composites Exhibit Tunable Stiffness and Power-Law Stress Relaxation | Biophysical Journal | 2018 | ISSN 1542-0086 |
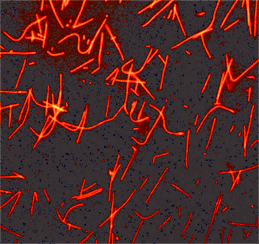
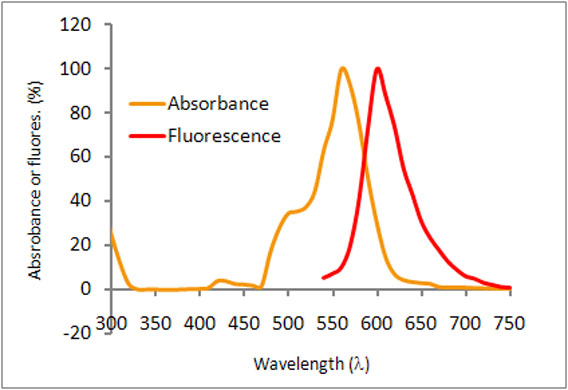
| Zhu, Yili et al. | An in vitro Microscopy-based Assay for Microtubule-binding and Microtubule-crosslinking by Budding Yeast Microtubule-associated Protein | Bio-Protocol | 2018 | ISSN 2331--8325 |
| Murray, John W. et al. | Reduction of organelle motility by removal of potassium and other solutes | PLoS ONE | 2017 | ISSN 1932-6203 |
| Shim, Albert et al. | Gliding Assay to Analyze Microtubule-based Motor Protein Dynamics | Bio-Protocol | 2017 | ISSN 2331--8325 |
| Arellano-Santoyo, Hugo et al. | A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity | Developmental Cell | 2017 | ISSN 1878-1551 |
| Reinemann, Dana N. et al. | Collective Force Regulation in Anti-parallel Microtubule Gliding by Dimeric Kif15 Kinesin Motors | Current Biology | 2017 | ISSN 0960-9822 |
| Britto, Mishan et al. | Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism | Proceedings of the National Academy of Sciences of the United States of America | 2016 | ISSN 1091-6490 |
| Shapira, Ofer et al. | Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain | Scientific Reports | 2016 | ISSN 2045-2322 |
| Zitouni, Sihem et al. | CDK1 Prevents Unscheduled PLK4-STIL Complex Assembly in Centriole Biogenesis | Current Biology | 2016 | ISSN 0960-9822 |
| Bartsch, Tobias F. et al. | Nanoscopic imaging of thick heterogeneous soft-matter structures in aqueous solution | Nature Communications | 2016 | ISSN 2041-1723 |
| Kim, Kyongwan et al. | Electric field-induced reversible trapping of microtubules along metallic glass microwire electrodes | Journal of Applied Physics | 2015 | ISSN 1089-7550 |
| Szyk, Agnieszka et al. | Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase | Cell | 2014 | ISSN 1097-4172 |
| Volkov, Vladimir A. et al. | Preparation of segmented microtubules to study motions driven by the disassembling microtubule ends | Journal of Visualized Experiments | 2014 | ISSN 1940-087X |
| Kitagawa, Mayumi et al. | Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis | Cell Reports | 2014 | ISSN 2211-1247 |
| Hawkins, Taviare L. et al. | Mechanical properties of doubly stabilized microtubule filaments | Biophysical Journal | 2013 | ISSN 0006-3495 |
| McVicker, Derrick P. et al. | The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of Tau to inhibit kinesin-mediated transport | Journal of Biological Chemistry | 2011 | ISSN 0021-9258 |
| Mori, Masashi et al. | Intracellular Transport by an Anchored Homogeneously Contracting F-Actin Meshwork | Current Biology | 2011 | ISSN 0960--9822 |
| Mukhopadhyay, Aparna et al. | Proteomic analysis of endocytic vesicles: Rab1a regulates motility of early endocytic vesicles | Journal of Cell Science | 2011 | ISSN 0021-9533 |