Post-Translational Modification Detection Techniques

Post-translational modifications (PTMs) like phosphorylation, acetylation, SUMOylation, ubiquitination, and others account for the vast increase in proteome complexity [1]. For any given protein, a variety of PTMs offer a way to facilitate rapid cellular changes by altering the structure and function of the protein. PTMs play a critical role in signal transduction, protein stability and turnover, protein-protein recognition and interaction, as well as spatial localization [2].
Due to the importance of PTMs in basic biology as well as in disease pathogenesis, there is a significant interest in identifying regulatory PTM mechanisms for every protein of interest (POI). Investigation of PTM modified POIs normally require an enrichment step as these modified proteins are relatively low in abundance. The majority of PTM detection methods have been developed in combination with enrichment strategies to provide the best opportunity to identify, validate, and study the function of regulatory PTMs for a POI [3,4]. However, there are a limited number of PTM modified proteins where site specific PTM modified antibodies have been developed; for example, EGFR pY1045. [5]. In these relatively rare cases enrichment steps may not be necessary. For all other situations see below for more information about commonly used assays for PTM detection.
Choosing the appropriate assay, method, and/or protocol will depend on the question you are trying to answer; ultimately, it is recommended to use a combination of these methods for identification, validation, and mechanistic characterization of a PTM for a POI.
Immunoprecipitation (IP) is a core technique that is utilized in several different PTM detection assays. Importantly, IP allows researchers to enrich for specific, low-abundance PTM modifications on a target POI. Enrichment is achieved through affinity-based purification technology. Antibodies (or specific binding molecules) attached to a solid support matrix (such as agarose resin) bind to the POI or PTM, while non-targeted proteins in the complex lysate are not captured and removed through wash steps. The enriched proteins are then removed off of the support matrix using elution buffers and isolated in a concentrated volume. The isolated population is analyzed by downstream methods like western blot or by mass spectrometry to determine if a POI is post-translationally modified.
IP is a critical step in the majority of PTM detection techniques; thus, having optimized, high-quality IP reagents will provide the best likelihood of obtaining meaningful results. These reagents include the appropriate lysis buffer system, antibody conjugated to affinity beads (to minimize antibody contamination), control beads, wash buffer, and elution buffer. See Figure 1 for the basic IP steps, and click here for detailed tips for successful IP enrichment.
Click here for a step-by-step video protocol in JoVE.
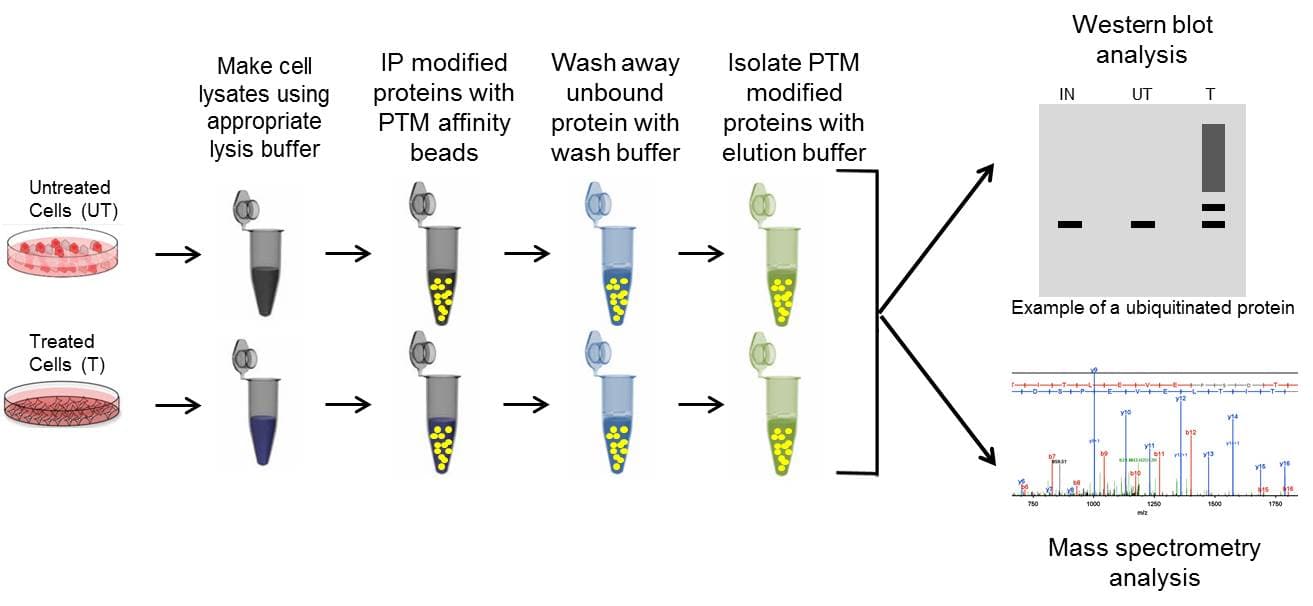
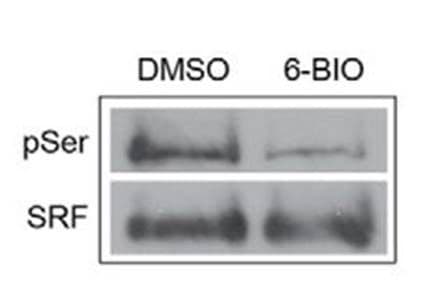
Protein specific IP utilizes an antibody against a POI to immunoprecipitate potentially all species of that protein. The enriched proteins are then separated by SDS-PAGE, transferred to a PVDF membrane, and analyzed via western blot, by probing with a target PTM antibody. Figure 2 provides example data utilizing POI-antibody specific IP, where phosho-SRF was detected using an SRF antibody for IP follwed by phospho serine antibody detection [6]. Target protein antibody IP has traditionally been the first approach used to determine if a POI is modified by a particular PTM, possibly due to the investigator's expertise with the POI specific antibody for western blot applications.
However, as Figure 1 shows, utilizing the antibody in an IP assay is quite different than its use in a western blot application. It is not surprizing that often times IP with a POI specific antibody fails, or requires significant optimization because it is performing a different function in an IP vs western blot application; therefore, as a starting point ensure that the POI specific antibody has been validated for IP. For initial discovery experiments, performing an IP with a PTM specific antibody may be an easier approach, where IP is performed with a PTM specific antibody and the POI specific antibody is used for western blot analysis. This approach only requires optimization of the IP step rather than having to optimize both the IP and the western steps which may be required in a proteinspecific IP.
Another point of consideration, is that previous research has shown that PTM modifications may block the antibody binding site on a POI; thus, false negatives are possible [7]. Either PTM specific IP or ovexpression IP may circumvent this problem, and should be considered if target antibody IP fails. See below for details on these two approaches.
Pros:
Cons:
PTM specific IP utilizes an antibody against a target PTM to immunoprecipitate potentially all proteins that have been modified by that PTM (dependent on the quality of the PTM affinity reagent). The enriched post-translationally modified population is then separated by SDS-PAGE analysis, transferred to a PVDF membrane, and analyzed via western blot, by probing with an antibody targeting a specific POI.
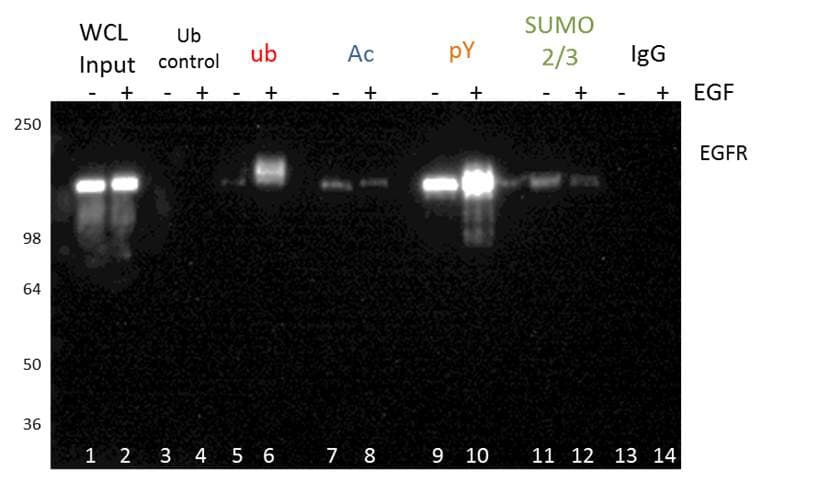
Figure 3 shows an example of PTM specific IP using tyrosine phosphorylation, ubiquitination, acetylation, and SUMOylation2/3 PTM affinity beads. The EGFR PTM profile of these four modifications was analyzed by probing with an EGFR antibody. This methodology is great for initial discovery of novel PTMs for any POI. PTM specific IP enrichment is also commonly used for bottom-up proteomics (see below for details), and may be benefical when investigating global PTM changes [8].
The PTM specific IP approach is particularly benfical as kits are available in this format, which allows the investigator to bypass issues that arise with protein specific IP. For example, in a kit format, the antibody is routinely conjugated to beads to minimize heavy and light chain leaching. The Signal-Seeker PTM detection kit in particular provides investigators with key inhibitors, optimized buffer systems, and filters to enhance isolation of modified proteins, and reduce the optimization time. A significant benefit of the Signal-Seeker kits is its ability to capture low-abundance, endogenous PTMs. This feature of the kit was highlighted in a recent publication investigating the clinically relevant PD-L1 protein [9].
Click here for a detailed protocol for PTM specific IP using Signal-Seeker™ PTM detection kits.
Pros:
Cons:
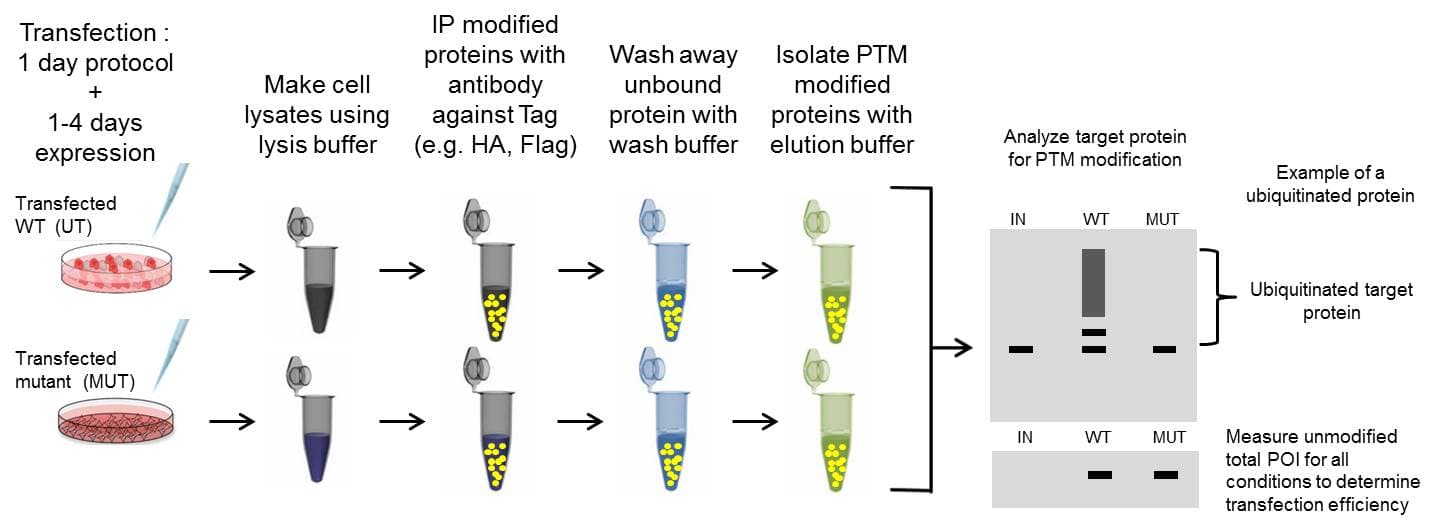
Overexpression IP is a well-established system where a plasmid of a tagged version of a POI is transfected into cells. This usually results in high expression levels of the tagged POI, which can be immunoprecipitated with a well-characterized antibody against the tag (ex. Flag). The increased expression and optimized antibody improves the chance of identifying the PTM modified POI. Overexpression IP assays are normally analyzed by western blot. The experimental procedure is outlined in Figure 4, but only summarizes the tranfection step as it can vary widely depending on methodolgy and cell type.
The overexpression system is particularly important when a PTM for a POI is challenging to study endogenously; however, any critical findings should be validated with endogenous protein for authenticity [10]. Still, a significant benefit of the overexpression method include the ability to create mutated forms of the target protein to determine which specific amino acids are modified by a specific PTM.
It is important to note overexpression IP assays are not effective for analyzing changes in the target protein's physiologic PTM state, as overexpression may mask results. Not only is expression artifical, but tags may also interfere with PTM regulation and protein interactions as well. When studying physiologic PTM changes to a target POI, immunprecipitating endogenous protein is recommended, which may be achieved using standard protein or PTM specific IP.
If overexpressing a protein without a tag, then IP should be performed with either a protein or PTM antibody IP. In many cases overexpression is performed because the POI specific antibody doesn't work. Utilizing a comprehensive IP detection system like the Signal-Seeker kit may provide the best enrichment strategy in these situations.
Pros:
Cons:
Nearly all Mass spectrometry analysis of PTMs are more effective when combined with IP strategies discussed above to enrich for a protein or PTM of interest. Rather than performing a western blot to determine if a specific protein is modified, the sample is analyzed using a mass spectrometer. The investigator can identify a spectrum of proteins modified by a PTM using bottom-up peptide-based PTM proteomics [3, 8, 11]. Mass spectrometry based PTM identification is a powerful tool, and is a valuable complement to conventional western blot PTM identification; especially for site specific identification.
Additional benefits of mass spectrometry are its potentially unbiased approach, and independence from antibodies for detection of PTM modified proteins. While mass spectrometry theoretically provides an unbiased snapshot of PTM modified proteins, it has several limitations which bias against certain types of proteins. Some of the technical challenges include protein abundance bias [12], and method sensitivity [13]. Fiest and Hummon provide a general review on instrumentation, sample preparation, decontamination, digestion strategies, fractionation approaches, and other considerations when utilizing bottom-up mass spectrometry [14].
There is no guarantee that a particular PTM of a POI can be identified using bottom-up mass spectrometry approaches. Of note, a given protein can be identified by multiple peptides within its sequence, but in order to identify a specific PTM for a protein, that particular peptide needs to be identified; obviously, this significantly lowers the chance for identification [15]. One strategy to enhancing the number of modified peptides being detected by the mass spectrometer is to perform a peptide enrichment using a PTM affinity reagent rather than protein enrichment using a PTM affinity reagent. This will decrease the number of unmodified peptides that are enriched. Utlimately, to improve the odds of identifying low abundane post-translationally modified proteins, using the highest quality enrichment tools is paramount [15]. The Signal-seeker affinity beads, which have been validated against commonly used reagents may be extremely beneficial for isolating a robust sample of high and low abundance PTM modified proteins (see acetyl-lysine tools white paper as an example).
A newer mass spectrometry technology involves analyzing the entire protein (top-down proteomics), and identifying all PTM modifications on the target POI [13]. This approach is known as top-down proteomics and involves IP with a target POI antibody. Unfortunately this technology is quite new so there are protein size limitations [13], a lack of access to the highly specialized proteomic tools, and a lack of available expertise like there is with the bottom-up approach.
All things considered, initial identification of a specific PTM modified protein using western blot approaches may be easier, require less time, and may be a good first step before utilizing mass spectrometry to address site-specific questions.
Pros:
Cons:
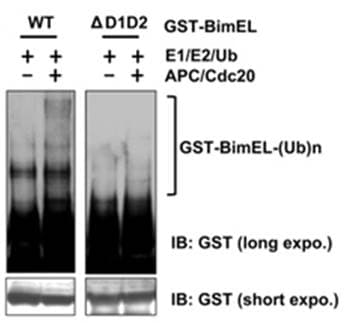
Biochemical assays utilize purified or in vitro translated versions of a target protein to determine if it can be modified by a specific PTM. The purified protein is added to a test tube with specific enzymes (e.g. E1, E2, E3 ubiquitin ligase) and the appropriate substrate (e.g. ubiquitin), co-factors, and energy sources. After incubation, the sample is then analyzed by western blot analysis. Figue 6 provides an example of in vitro ubiquitination of the apoptosis protein BimEL [16].
It is important to note that in vitro biochemical analysis is not available for all types of PTMs; however, in vitro PTM analysis is routinely performed to validate phosphorylation [17], ubiquitination [9], SUMOylation [18] and other PTM modifications. A limiting step in performing in vitro biochemical assays is obtaining purified versions of the POI [19].
Pros:
Cons:
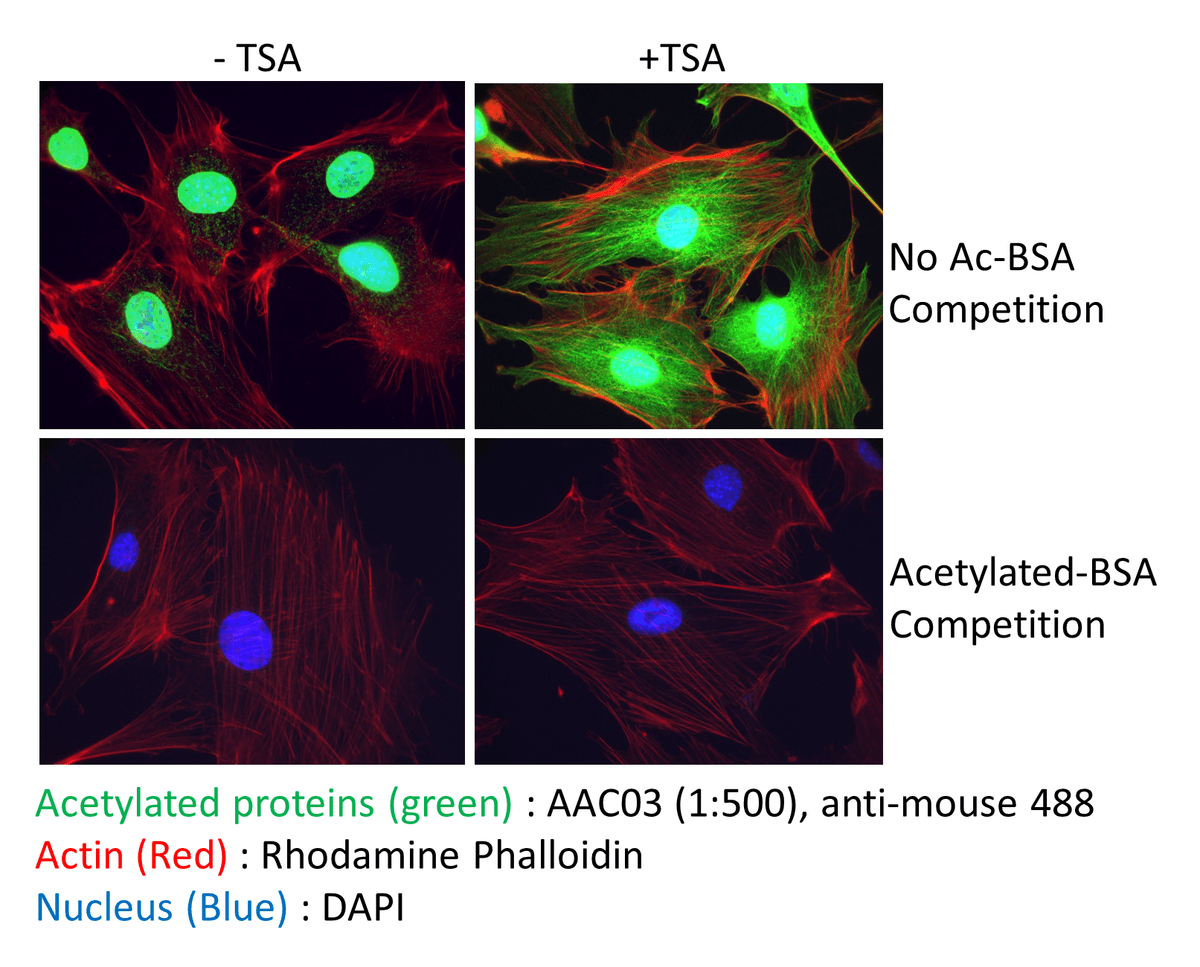
Immunofluorescence techniques may be a fruitful approach to studying global changes in a PTM profile in tissues or cells. In particular identifying global, and spatial changes in response to drug treament (e.g. HDAC inhibitors) or genetic knockout is achievable with this method. Figure 7 shows example data utilizing Signal-Seeker acetyl lysine antibodies to probe for total acetyl lysine changes. Several Signal-Seeker PTM antibodies have been validated for IF applications.
The identification of target specific PTM modifications are not possible by this method. However, this approach may be applicable as a biological readout; thus, may be a useful tool as an indicator for development [20,21] or disease progression [22,23].
Pros:
Cons:
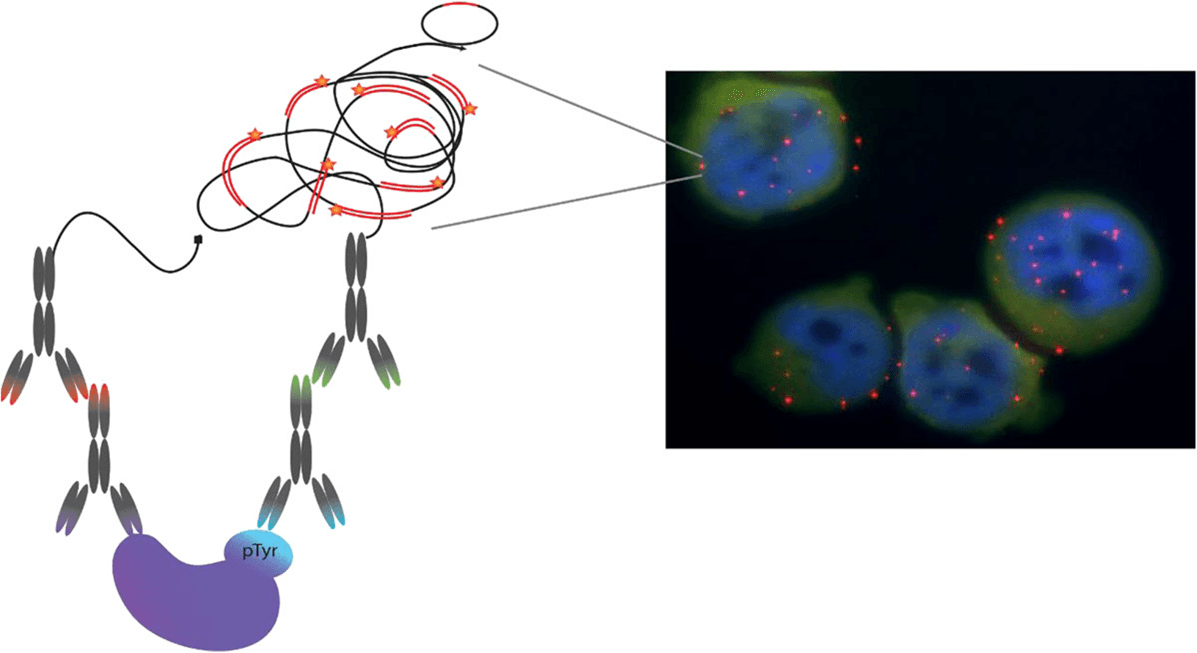
Proximity ligation assay (PLA) is a novel immunoassay technology that can be used to study protein interactions and PTMs. PLA is unique in its ability to identify PTMs of a specific POI in fixed tissues and cells[25]. The technology utilizes an amplification process, which allows detection of low abundance molecules and modifications [26]. Figure 8 provides a schematic of the PLA principle as well as example data generated using this approach [24].
PLA principle [24-26]:The principle of PLA-PTM works by utilizing two antibodies (preferably raised in different species) one against a PTM of interest and one for the target protein of interest. The initial steps in the procedure are similar to standard immunofluorescence staining where the primary antibodies bind the protein of interest and the PTM of interest.
Secondary antibodies are added that recognize the two primary antibodies. The difference with PLA is that these secondary antibodies have short DNA strands covalently attached to them (these antibody-DNA complexes are called PLA probes). If the two PLA probes are in close enough proximity, presumably because the two antibodies are binding to a single protein that has been PTM modified, they will form circularized DNA.
The next step requires the addition of polymerase, and amplification of circularized DNA. Once amplification is complete, fluorescently labeled complementary DNA probes are added. Due to the significant DNA amplification, which can be up to several hundredfold, the fluorescent signal from very few molecules will be visible by microscopy.
Pros:
Cons:
How to study PTMs: A Guide for new PTM Investigators
Bioscience Reports Article: Simple toolsets to identify endogenou PTMs for a target protein
Neoplasia Article: Identifying Regulatory Posttranslational Modifications of PD-L1
JoVE Article: Visual Protocol For Investigating Post- translational Modifications of Target Proteins