+3
Loading...
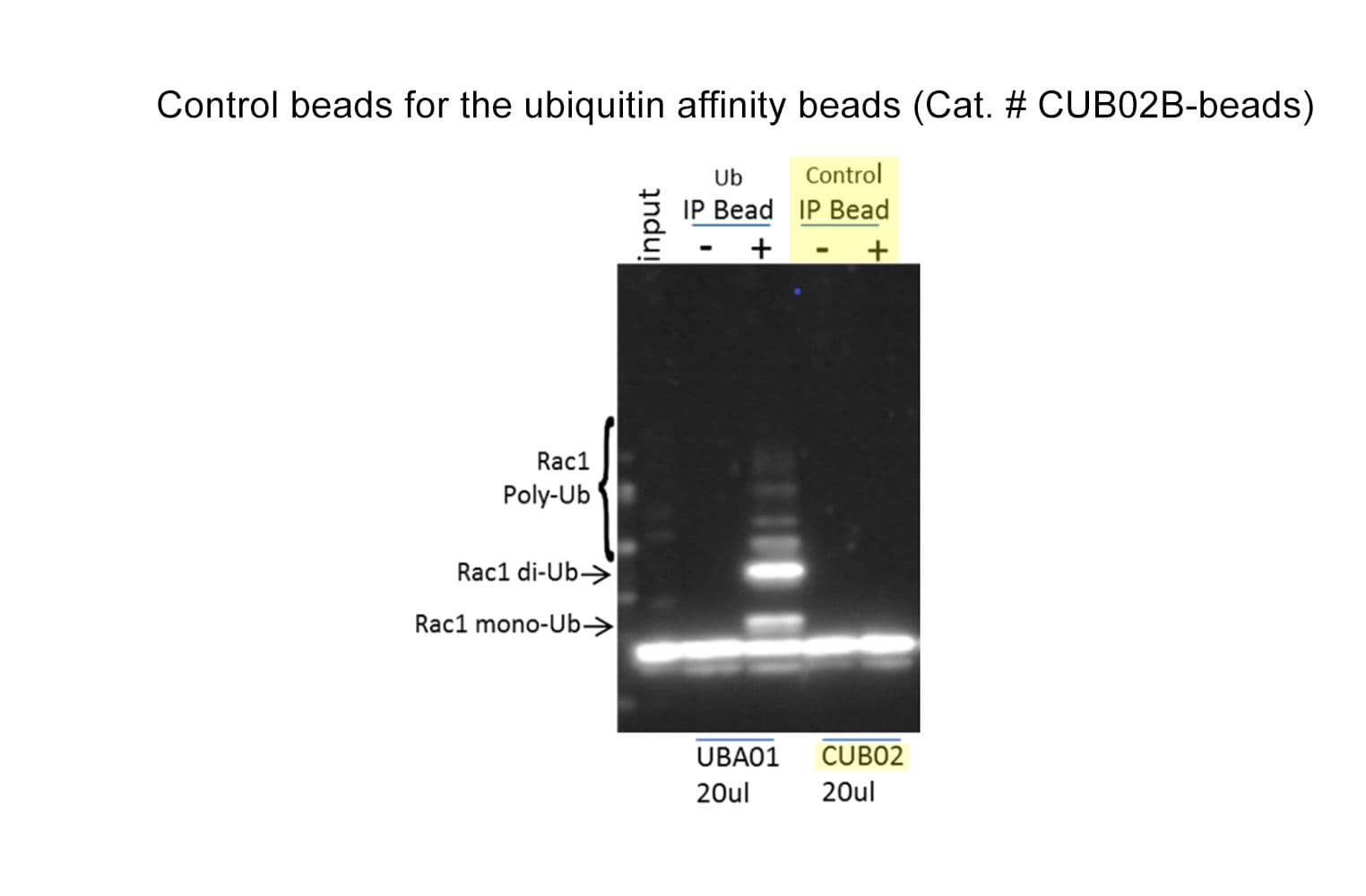
When performing ubiquitinated protein enrichment, it is important to include a control bead that is as similar to the enrichment beads as possible, but cannot bind ubiquitinated proteins. The CUB02B-beads contain two point mutations in the ubiquitin affinity binding domains of the ubiquitin enrichment beads Cat # UBA01B-beads that result in loss of Ub binding and offer an excellent control bead for Ub enrichment.
CUB02B-beads comprise non-Ub binding TUBE protein (due to point mutations) that has been co-valently attached to agarose beads. The point mutation
Key characteristics
Cat. #CUB02B-beads