Actin protein (rhodamine): rabbit skeletal muscle
Product Uses Include
- In vivo actin polymerization studies (microinjection into muscle cells)
- In vitro motility studies using fluorescent F-actin and muscle myosins
Material
Purified rabbit muscle actin (Cat. # AKL99) has been modified to contain covalently linked rhodamine at random surface lysine residues. An activated ester of rhodamine is used to label the protein. The labeling stoichiometry has been determined to be 1-2 dyes per actin monomer. Rhodamine labeled rabbit muscle actin has an approximate molecular weight of 43 kDa, and is supplied as a pink lyophilized powder. The lyophilized protein is stable for 6 months when stored desiccated to <10% humidity at 4°C. The protein should be reconstituted to 10 mg/ml with distilled water, it will then be in the following buffer: 5 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 5% sucrose, and 1% dextran.
Rhodamine actin from a non-muscle source is also available (Cat. # APHR).
Purity
Protein purity is determined by scanning densitometry of Coomassie Blue stained protein on a 12% polyacrylamide gel. AR05 rhodamine muscle actin was found to be >99% pure (see Figure 1).
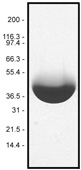
Figure 1. A 100 µg sample of rhodamine muscle actin (molecular weight approx. 43 kDa) was separated by electrophoresis in a 12% SDS-PAGE system, and stained with Coomassie Blue. Protein quantitation was performed with the Precision Red Protein Assay Reagent (Cat. # ADV02).
Biological activity
The biological activity of rhodamine muscle actin can be determined by its ability to efficiently polymerize into filaments in vitro and separate from unpolymerized components in a spin down assay. Stringent quality control ensures that 90% of the labeled muscle actin can polymerized in this assay, which is similar to the unlabeled product (Cat. # AKL99)
For product Datasheets and MSDSs please click on the PDF links below. For additional information, click on the FAQs tab above or contact our Technical Support department at tservice@cytoskeleton.com
Question 1: What is the best way to store actin proteins to insure maximum stability and shelf-life?
Answer 1: Cytoskeleton provides all actin proteins as lyophilized powders so that they can be shipped at room temperature. Upon receipt, the lyophilized powders should be stored at 4°C in a sealed container with desiccant. It is important to monitor the freshness of the desiccant and insure that it continues to absorb moisture to protect the lyophilized actins. With proper storage, the lyophilized actins are guaranteed to be stable for 6 months. Alternatively, actins can be immediately resuspended at the concentration recommended, aliquoted, snap-frozen in liquid nitrogen and stored at -70°C. When thawing frozen aliquots, it is important to thaw rapidly in a room temperature water bath.
Question 2: What is the best way to store F-actin after polymerizing?
Answer 2: G-actin is stable for two days at 4°C and requires a divalent cation, pH 6.5 - 8.0 and ATP for stability. F-actin is stable and can be stored at 4°C for 1-2 weeks. F-actin requires ATP (0.2 mM) and Mg2+ (2 mM) for stability and is unstable below pH 6.5 and above pH 8.5. F-actin is not stable to freezing. F-actin can be transferred to a variety of buffers (e.g. HEPES, phosphate, etc) without detrimental effects. We recommend the addition of antibacterial agents such as 100 μg/ml ampicillin and 10 μg/ml chloramphenicol when storing F-actin at 4°C.
Question 3: Filters for visualizing rhodamine signal?
Answer 3: The excitation filter should be set at 535 nm and the emission filter at 585 nm.
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com














