+3
Kit contents (96 assays: strip-wells allow single or multiple assays per run)
Equipment & materials required
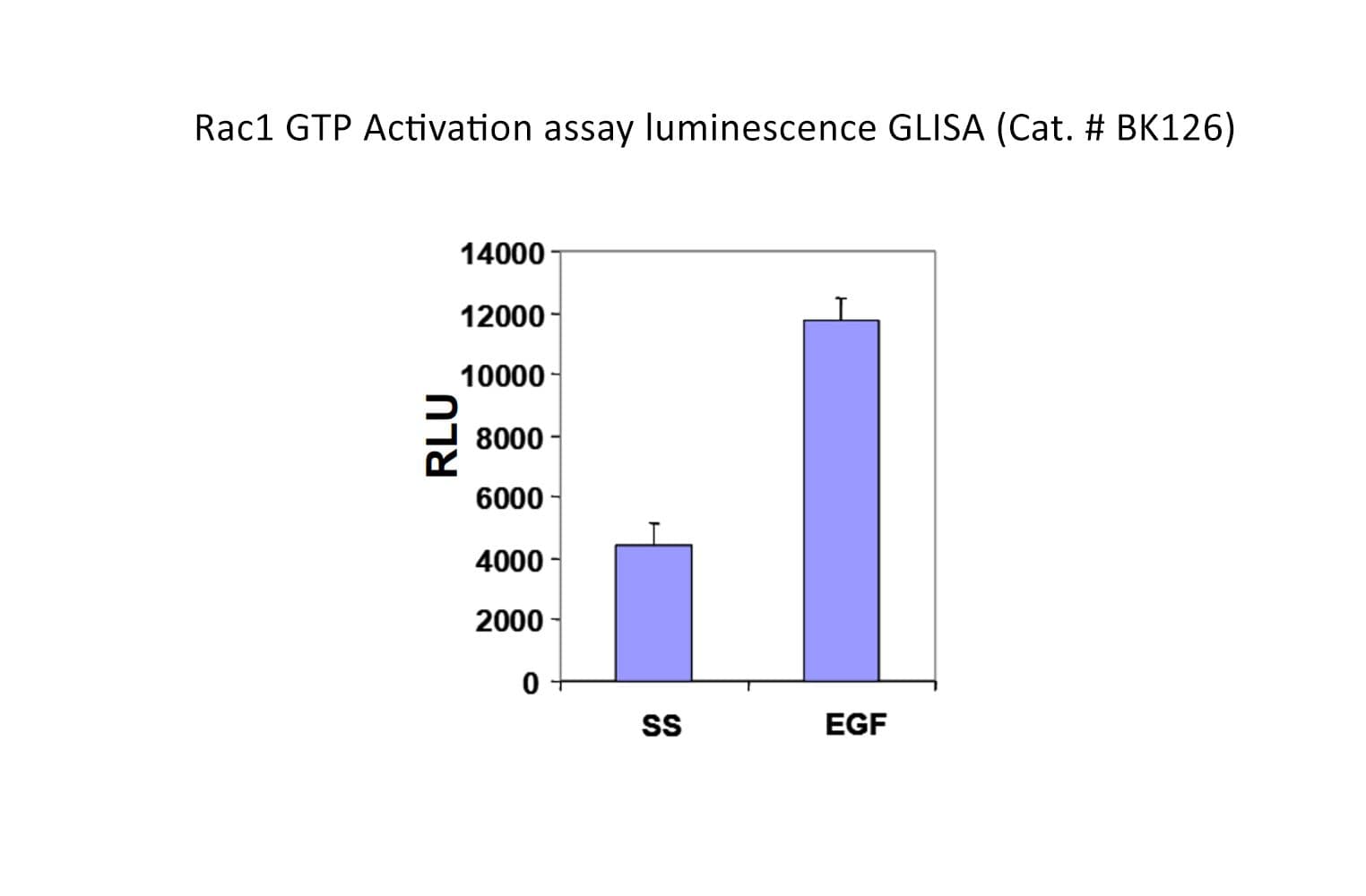
The Rac1 G-LISA® assay isolates active Rac/Cdc42 (GTP-bound form) using the Rac/Cdc42-binding domain (PBD) of an effector protein coupled to a 96-well strip plate. The bound Rac1 is then detected and quantified using a luminescent ELISA-based method and a Rac1-specific antibody.
Key characteristics
Cat. #BK126