About ADP-ribosylation factor (Arf) small G-proteins
The mammalian ADP-ribosylation factor (Arf) family of the Ras superfamily of GTPases consists of six Arf isoforms (Arf1-Arf6). The Arf GTPases were originally named for their ability to stimulate cholera toxin-mediated ADP ribosylation of the Gsα subunit utilized by many G-protein coupled receptors1. These GTPases are grouped into three classes based on their size and amino acid similarity: class I (Arf1, Arf2, Arf3), class II (Arf4 and Arf5), and class III (Arf6)2. Arf2 is not found in humans, while Arf1 and Arf6 are the focus of most Arf research efforts. Arf1 and Arf6 have distinct but similar functions in protein and membrane vesicle trafficking in the cell2,3, with roles in endosome and vesicle fusion as well as clathrin-mediated endocytosis.
Role of Arf1 in regulating actin assembly during vesicle formation at the Golgi.
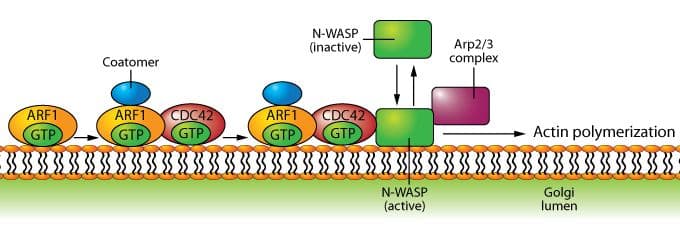
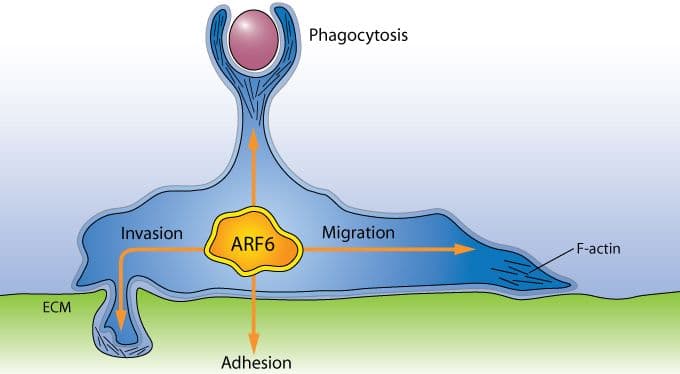
Historically, Arf1 has been viewed as having a primary role in the development and maturation of coated vesicles that transport proteins from the endoplasmic reticulum to the Golgi apparatus (see Arf1 figure), but recent data have also implicated Arf1 in the transport of proteins from the Golgi membrane to the plasma membrane4, as well as phagocytosis5. Arf6 is localized at the plasma membrane and endocytic compartments where the protein plays a significant role in endocytic membrane trafficking and localized actin dynamics, the latter involving cross-talk with Rho family GTPases3 (see Arf6 figure).