+3
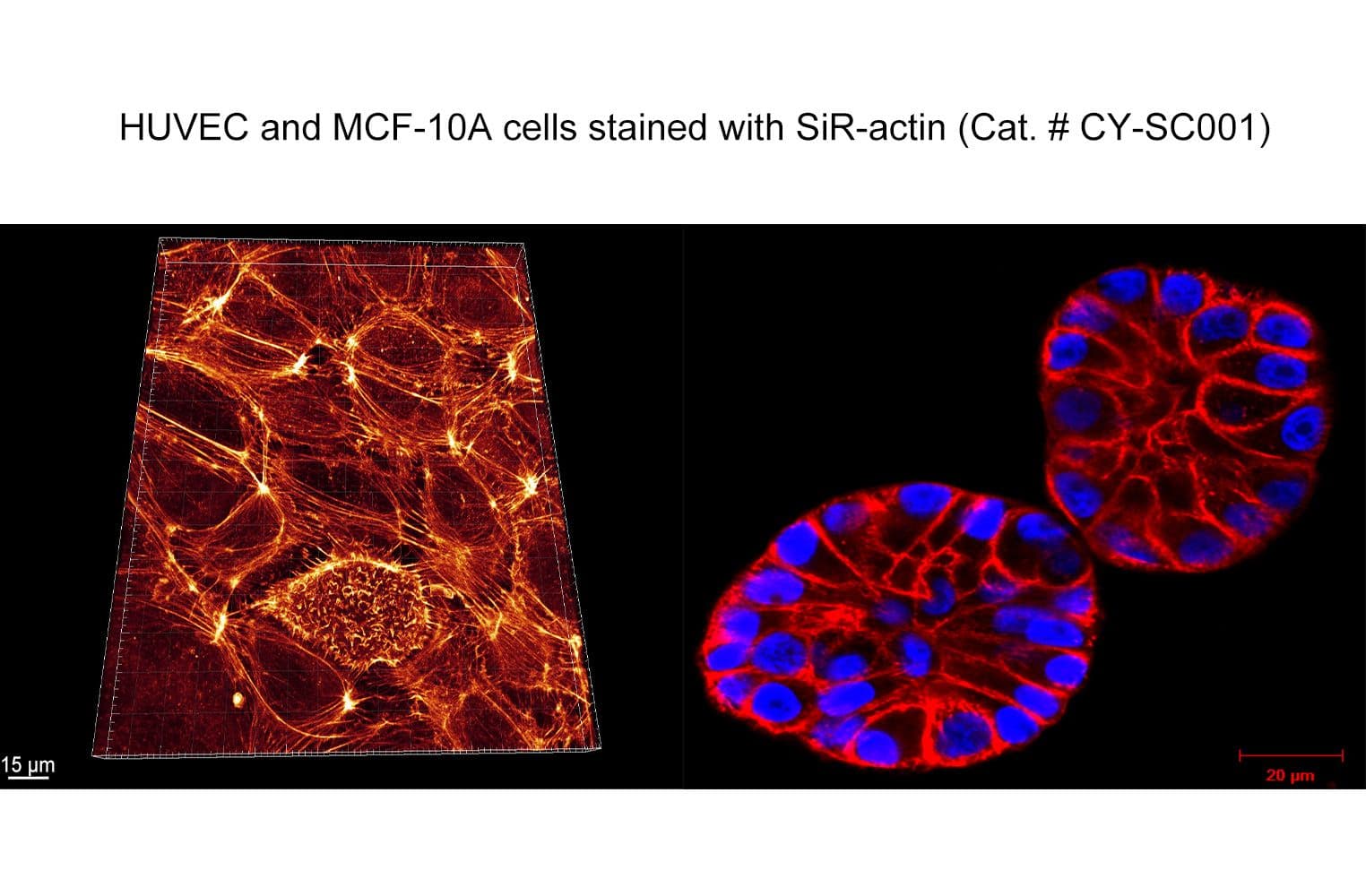
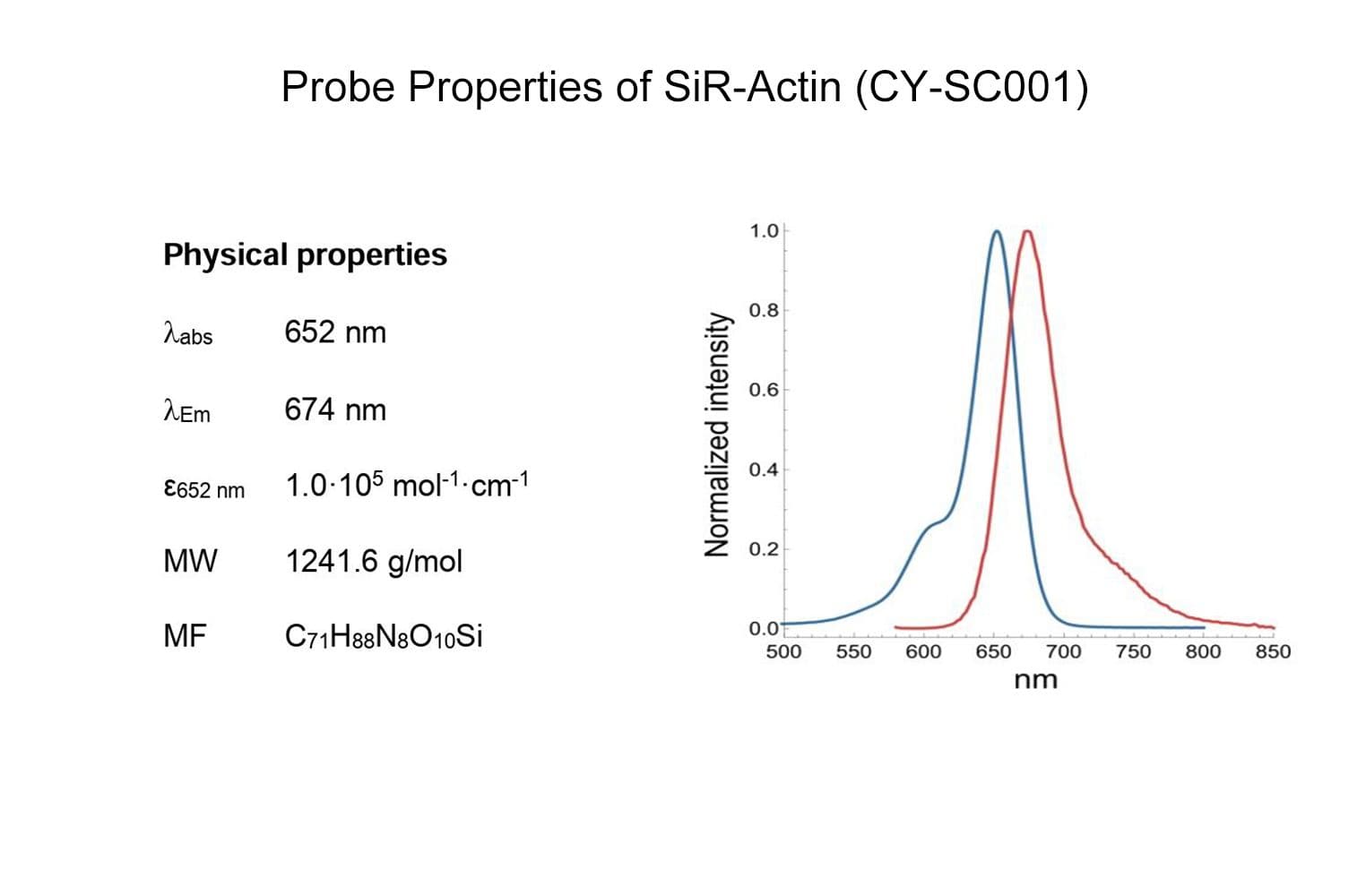
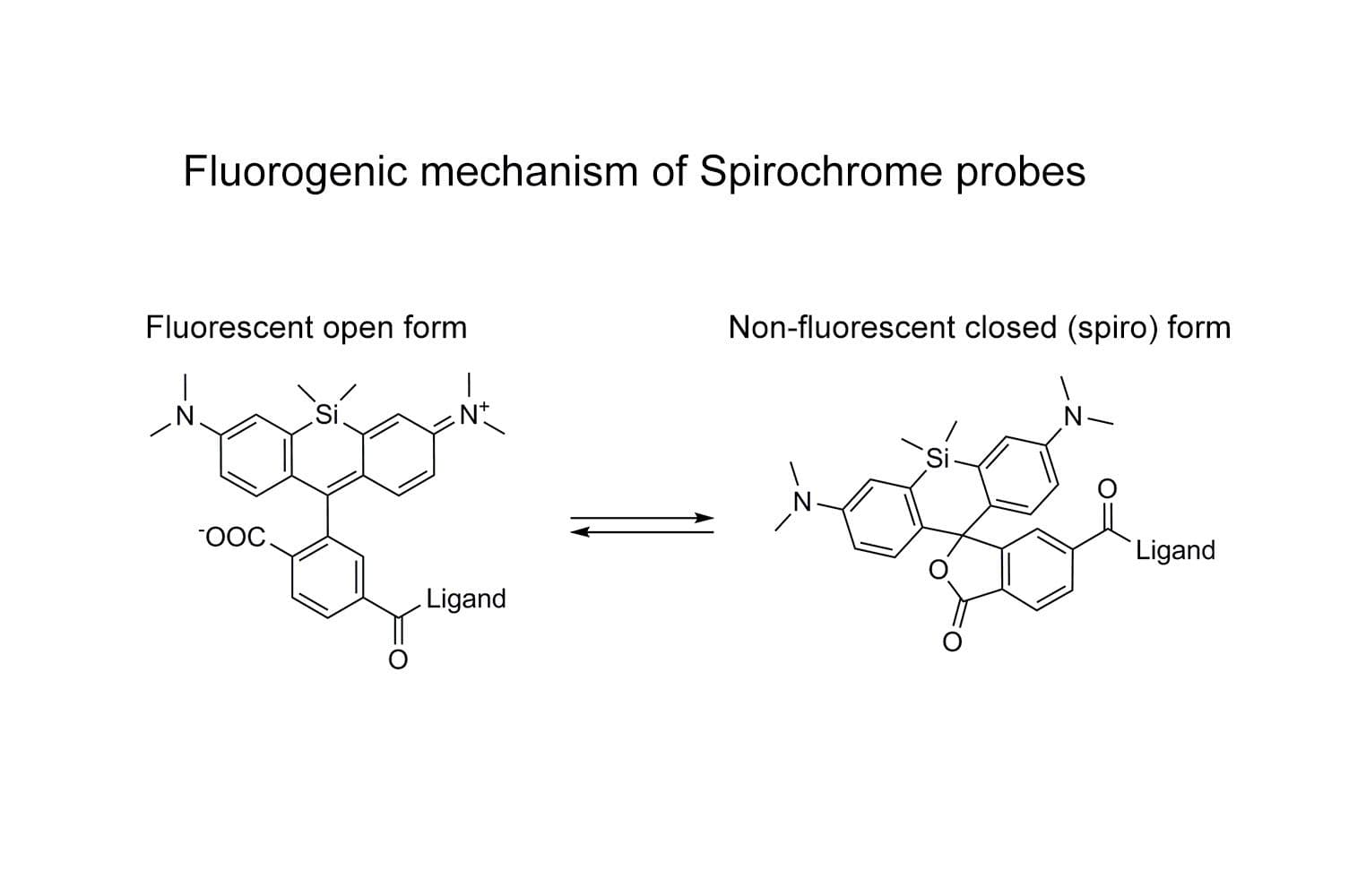
SiR-actin is based on the fluorophore silicon rhodamine (SiR) and the actin-binding natural product jasplakinolide. SiR-actin allows the labelling of F-actin in live cells with high specificity and low background. Its superior fluorogenic properties make this the probe of choice for low-background imaging of dynamic actin filaments. FastAct™ /FastAct™_X probes are recommended for imaging highly dynamic actin processes.
Key features
The biological activity of CY-SC001 is assessed by the ability of the probe to efficiently label actin filaments in live cell culture. After a wash step, cell staining is visible for several hours.
Cat. #CY-SC001