Microtubule Binding Protein Spin-Down Assay Biochem Kit
Product Uses Include
- To determine whether a protein or compound binds to microtubules.
- To test various deletion mutants for their ability to still bind microtubules, then calculate their affinity for microtubules, and hence identify the microtubule binding site.
- In the presence of a non-hydrolyzable analog of ATP (kinesins) or GTP (dynamin) extract these from cells or show their binding affinity when mutated.
Introduction
This assay allows the identification of proteins that will bind to microtubules (MTs) in vitro. The assay relies on the fact that MTs will pellet when centrifuged at 100,000 x g. Therefore, any protein that is associated with the MTs will pellet with them during centrifugation. A simple SDS-PAGE analysis of the supernatant versus pellet fraction will identify if a protein is able to associate with MTs. The assay description given in this manual is for recombinantly expressed “test” proteins, however, the assay can be adapted for cell lysates or in vitro translation products.
It should be noted that in vivo confirmation of MT association should be obtained in order to confirm that the protein can be classified as a MAP. This association need not occur throughout the whole cell cycle and may even be developmentally regulated, indeed transient association of MAPs with microtubules is the norm rather than the exception.
Kit contents
The kit contains sufficient materials for 30-100 assays depending on assay volume. The following reagents are included:
- Tubulin, >99% pure, (Cat. # T240)
- Microtubule associated protein fraction (positive control) (Cat. # MAPF)
- Bovine serum albumin (BSA) protein (negative control)
- General tubulin buffer (Cat. # BST01)
- Tubulin glycerol buffer (Cat. # BST05)
- GTP (Cat. # BST06)
- Microtubule resuspension buffer
- Paclitaxel (Cat. # TXD01)
- DMSO
- Salt-extraction buffer
- Manual with detailed protocols and extensive troubleshooting guide
Equipment needed
- TCA solution 50% (w/v) for protein precipitation if necessary.
- Centrifugation set-up capable of 100,000 x g at 4°C and 24°C, 50 -200 µl volume capacity.
- SDS-PAGE system.
- Detection system for protein of interest (coomassie is good for purified proteins, Western blot or silver stain for less pure or low abundance test proteins).
- Gel scanner for densitometric determinations.
Example results
The microtubule binding spin-down assay was tested by combining MTs with the a microtubule associated protein fraction (Cat. # MAPF) or with BSA. As expected, MAPFs co-sediment with the microtubules while BSA does not (Fig. 1)
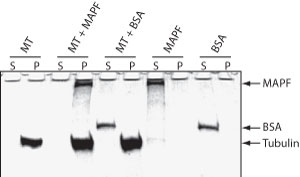
Figure 1. MAP Spin-Down Assay using BK029. Microtubules were mixed with buffer (MT), MAP proteins (MT + MAPF) or BSA (MT +BSA) and MTs were pelleted by centrifugation at 100,000 x g. Supernatant (S) and Pellet (P) fractions were examined by SDS-PAGE. MAP proteins, but not BSA, bind to MTs and co-precipitate. MAP (MAPF) or BSA (BSA) proteins alone do not pellet.
For product Datasheets and MSDSs please click on the PDF links below. For additional information, click on the FAQs tab above or contact our Technical Support department at tservice@cytoskeleton.com
Question 1: Can cell lysates be used with this kit as the source of a test protein?
Answer 1: Yes, cell lysates can be used as the source of the test protein for examining binding between tubulin monomers, polymers and the protein of interest. However, Cytoskeleton does not recommend this as the purity and concentration of the protein will often be too low to interact with tubulin monomers and polymers. Also, the lysates will contain additional accessory proteins and multiple phosphatases and proteases that can interfere or alter the interactions between tubulin and the test protein. We recommend consulting these citations which used cell lysates with this kit: Monzo et al., 2005. Clues to CD2-associated protein involvement in cytokinesis. Mol. Biol. Cell. 16, 2891–2902; Moshnikova et al., 2008. Interaction of the growth and tumour suppressor NORE1A with microtubules is not required for its growth-suppressive function. BMC Research Notes. 1, 13. We also recommend the following modifications:
Use 4 x 15 cm plates of cells. Wash and scrape cells with a PIPES-based buffer at 4°C. Use 3 mls of the following buffer: 20 mM PIPES, pH 7.0, 2 mM MgCl2, 1 mM EGTA, 1 mM GTP, 0.5 mM fresh PMSF (a protease inhibitor that has only 30 min half-life in aqueous solutions), and 1:100 dilution of 100X protease inhibitor cocktail (Cat. # PIC02). Collect the scraped cells and lyse the cells with sonication at 4°C with 5 pulses on medium for 15 secs each with a 1 min cool down in between each sonication burst. Then centrifµge the lysate at 14,000 x g in a microfµge at 4°C for 20 min. Measure the protein in the supernatant and proceed by mixing 2 to 5 mg/ml of extract with 0.5 mg/ml microtubules, as described in the kit.
Question 2: What is an appropriate buffer for the test protein?
Answer 2: The test protein must be in an appropriate buffer to allow attachment to microtubules. Conditions which do not favor microtubule binding are: high salt (e.g. >50 mM Buffer or NaCl), high or low pH (i.e. pH of <6.0 or >9.0) and high calcium (greater than 0.1 mM calcium will depolymerize microtubules). EGTA is included at 1mM to chelate excess calcium.
It is often advisable to include 5.0 mM Mg2+ in the buffer. Appropriate buffers include: HEPES, PIPES and MES, all at 20 mM. In some cases, the presence of ATP or GTP in the reaction may prevent MAP/microtubule association (e.g., kinesin or dynamin, respectively).
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com




