Upon translation, proteins are trafficked to their proper subcellular location so that they can perform their physiological functions. One mechanism that mediates correct protein localization and function is post-translational modifications (PTMs). PTMs include protease cleavage, protein folding, and the attachment of molecules such as biochemical functional groups (e.g., acetate or phosphate) or small proteins (e.g., ubiquitin or SUMO [Small Ubiquitin-like Modifier]). In this way, PTMs provide cells with a second level of regulation of protein function, signaling, and fate1,2. PTM-mediated regulation involves alteration of protein binding motifs, thereby affecting the binding with other proteins, nucleic acids, and various small molecules. Enzyme-mediated reversible PTMs offer an especially powerful modulatory tool that enable proteins to perform multiple tasks systematically while the protein remains under precise regulatory control1,2.

SUMO proteins are 12 kDa proteins3 discovered 18 years ago by four independent research groups4-8 and since then, their regulation of cellular processes has been heavily investigated9,10. SUMO proteins are highly conserved from yeast to mammalian cells11. In budding yeast (Saccharomyces cerevisiae), only one SUMO protein (Smt3) exists, but in vertebrate, three major SUMO isoforms (SUMO-1, SUMO-2, and SUMO-3) exist, expressed in all tissue12. Because mature SUMO-2 and SUMO-3 are 97% identical in amino acid sequence (48% identity with SUMO-1) and no functional difference between them has been identified, they are collectively called SUMO-2/313. SUMO proteins (SUMO-1 vs SUMO-2/3) show distinct subcellular localization and the expression level of SUMO-2/3 is higher than that of SUMO-114-16. While only a single SUMO-1 is ligased to target proteins, multiple SUMO-2/3s form poly-SUMO-2/3 chains by attaching SUMO to a conserved lysine residue and this chain is terminated by SUMO-1 ligation17,18.
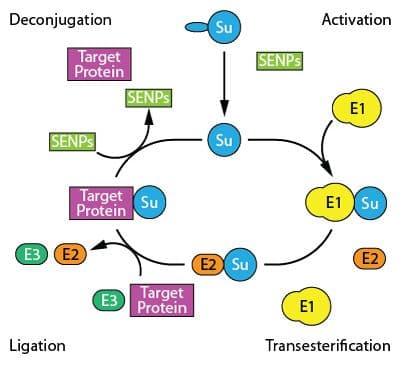
SUMOylation, the linkage of SUMOs to target proteins through isopeptide bonds, is orchestrated by a series of enzymatic reactions19 (Fig. 1). Processing enzymes (SENPs) cleave the C-terminus of newly synthesized SUMOs and the heterodimeric E1 SUMO activating enzyme (SAE1/2 subunits) activates SUMOs by the covalent attachment of SUMO's exposed di-glycine residue to SAE2's conserved cysteine by a thioester linkage19,20. The activated SUMOs are transferred to the sole E2 conjugating enzyme (Ubc9). Here, the SUMOs can be ligased to target proteins through two different pathways. First, the SUMO-Ubc9 complex binds to the target protein through a SUMO-interacting motif (SIM) and Ubc9 catalyzes isopeptide bonds between the C-terminus of SUMO and the epsilon amino group of lysines in the target protein, a process that is E3 enzyme independent22. The second mechanism involves various E3 ligases such as PIAS family members23, RanBP224, or Pc225 targeting preferential proteins24-28. Once SUMO-Ubc9 forms a complex with the E3 ligase, the E3 ligase binds to the target proteins and facilitates the SUMO ligation to target proteins29. Many SUMOylation reactions occur on a consensus motif, Ψ-K-x-D/E (Ψ; hydrophobic residue, K; lysine, x; any amino acid and D/E; aspartic acid/glutamic acid)30, but SUMOylation on non-canonical consensus motifs has been reported31,32.
Functionally, SUMOylation targets multiple proteins (Fig. 2), including Rac1 GTPase and the two cytoskeletal building block proteins actin and tubulin33,34. SUMOylation of Rac1, a Rho family GTPase, is mediated by the SUMO E3 ligase, PIAS3, and is necessary for increases in Rac1 activity in response to various growth factors and the ensuing Rac1-mediated lamellipodia/membrane ruffle formation which underlies cell migration and invasion34. The authors suggest that SUMOylation is not required for activation, but instead extends the time that Rac1 is active. Rac1 SUMOylation occurs under both in vitro and in vivo conditions and while both GTP-bound and GDP-bound Rac1 are SUMOylated, the active form of Rac1 is preferentially targeted34. Mass spectrometry revealed that 95% of in vitro Rac1 SUMOylation occurs on lysines 188, 183 and 184, or 186, residues within the C-terminus polybasic region34. These same lysine residues are also targeted for SUMOylation under in vivo conditions34. Upstream of Rho family GTPases is the cytoskeletal protein actin. In vitro SUMOylation of actin by both SUMO2 and SUMO3 is involved in the nuclear localization of actin19,35,36 with both SUMO2 and SUMO3 modifying nuclear actin through an interaction involving amino acids K68 and K28436.
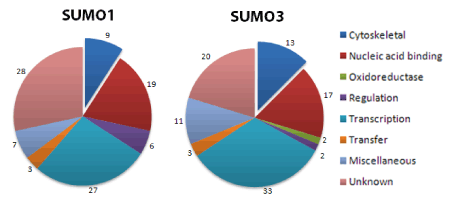
The other major cytoskeletal protein targeted by SUMOs is tubulin. SUMOylation of alpha-tubulin has been confirmed in vitro, while beta-tubulin has been identified as a putative SUMO-1 and SUMO-3 target19. The authors19 posit that SUMOylation of tubulin may enable the sequestering of other SUMOylated proteins for the purpose of assembling multimeric protein complexes.
In summary, SUMO-mediated regulation of protein localization and function is not only complex but well-positioned as a major regulator of cellular architecture and transport. To better understand how SUMOylation regulates proteins, Cytoskeleton, Inc. is focusing on the development of high-quality antibodies which are highly specific to PTMs.
Related Products & Resources
Select Post-translational Modified Products
Anti-Acetyl Lysine Mouse Monoclonal Antibody (Cat. # AAC01)
Anti-Phosphotyrosine Mouse Monoclonal Antibody (Cat. # APY03)
NEW! Anti-SUMO 2/3 Mouse Monoclonal Antibody (Cat. # ASM23)
NEW! Anti-SUMO 2/3 Mouse Monoclonal Antibody (Cat. # ASM24)
Anti-Ubiquitin Mouse Monoclonal Antibody (Cat. # AUB01)
References
- Yang X.J. 2005. Multisite protein modification and intramolecular signaling. Oncogene. 24, 1653-1662.
- Flotho A. & Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357-385.
- Bayer P. et al. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275-286.
- Boddy M.N. et al. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 13, 971-982.
- Shen Z. et al. 1996. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics. 36, 271-279.
- Okura T. et al. 1996. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 157, 4277-4281.
- Matunis M.J. et al. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell. Biol. 135, 1457-1470.
- Mahajan R. et al. 1997. A small ubiquitin-related polypeptide involved in targeting Ran-GAP1 to nuclear pore complex protein RanBP2. Cell. 88, 97-107
- Gareau J.R. & Lima C.D. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861-871.
- Jentsch S. & Psakhye I. 2013. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47, 167-186.
- Chen A. et al. 1998. Characterization of mouse ubiquitin-like SMT3A and SMT3B cDNAs and gene/pseudogenes. Biochem. Mol. Biol. Int. 46, 1161-1174.
- Huang W.C. et al. 2004. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 271, 4114-4122.
- Kamitani T. et al. 1998. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 273, 11349-11353.
- Saitoh H. & Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252-6258.
- Manza L.L. et al. 2004. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 17, 1706-1715.
- Ayaydin F. & Dasso M. 2004. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 15, 5208-5218.
- Bohren K.M. et al. 2004. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 279, 27233-27238.
- Tatham M.H. et al. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368-35374.
- Rosas-Acosta G. et al. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics. 4, 56-72.
- Johnson E.S. et al. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509-5519.
- Lois L.M. & Lima C.D. 2005. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439-451.
- Melchior F. 2000. SUMO--nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16, 591-626.
- Johnson E.S. & Gupta A.A. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 106, 735-744.
- Pichler A. et al. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108, 109-120.
- Kagey M.H. et al. 2003. The polycomb protein Pc2 is a SUMO E3. Cell. 113, 127-137.
- Schmidt D. & Muller S. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U.S.A. 99, 2872-2877.
- Ungureanu D. et al. 2003. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 102, 3311-3313.
- Sachdev S. et al. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088-3103.
- Lin D. et al. 2002. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277, 21740-21748.
- Hay R.T. 2005. SUMO: a history of modification. Mol. Cell. 18, 1-12.
- Kamitani T. et al. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273, 26675-26682.
- Rangasamy D. et al. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275, 37999-38004.
- Panse V.G. et al. 2004. A proteome-wide approach identifies sumolyated substrate proteins in yeast. J. Biol. Chem. 279, 41346–41351.
- Castillo-Lluva S. et al. 2010. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 12, 1078-1085.
- Vertegaal A.C. et al. 2004. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279, 33791-33798.
- Hofmann W.A. et al. 2009. SUMOylation of nuclear actin. J. Cell Biol. 186, 193-200.