How to study PTMs: A Guide for new PTM Investigators

Post-translational modifications (PTMs) like phosphorylation, acetylation, SUMOylation, ubiquitination, and others account for the vast increase in proteome complexity [1]. For any given protein, a variety of PTMs offer a way to facilitate rapid cellular changes by altering the structure and function of the protein. PTMs play a critical role in signal transduction, protein stability and turnover, protein-protein recognition and interaction, as well as spatial localization [2]. Due to the importance of PTMs in basic biology as well as in disease pathogenesis, there is a significant interest in identifying regulatory PTM mechanisms for every protein of interest (POI).
Key Points:
-- PTMs function as regulatory mechanisms for target proteins
-- PTMs are often transient
-- Due to its regulatory nature only a small fraction of a target protein may be modified by a target PTM
-- Enrichment strategies may be necessary to study a specific PTM for a target protein
Choosing the appropriate assay, method, and/or protocol will depend on the question you are trying to answer; ultimately, it is recommended to use a combination of these methods for identification, validation, and mechanistic characterization of a PTM for a target protein. Here we provide a general overview of immunoprecipitation based PTM enrichment, as well as important technical tips. For additional information on alternative methodologies and assays used to investigate PTMs click here to see our PTM Detection Techniques Page.
Immunoprecipitation based techniques
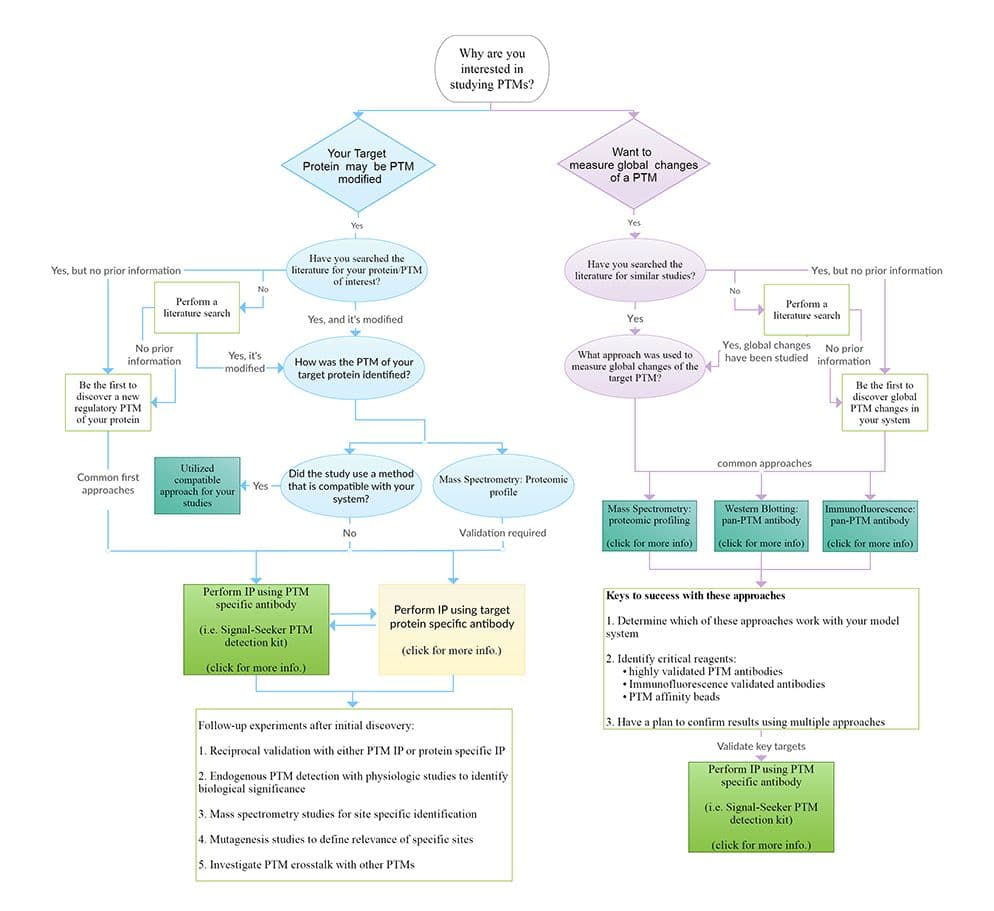
Immunoprecipitation (IP) is a core technique that is utilized in several different PTM detection assays. Importantly, IP allows researchers to enrich for specific, low-abundance PTM modifications on a target POI. Enrichment is achieved through affinity-based purification technology.
Antibodies (or specific binding molecules) attached to a solid support matrix (such as agarose resin) bind to the POI or PTM, while non-targeted proteins in the complex lysate are not captured and removed through wash steps. The enriched proteins are then removed off of the support matrix using elution buffers and isolated in a concentrated volume. The isolated population is analyzed by downstream methods like western blot or by mass spectrometry to determine if a POI is post-translationally modified. See Figure 1 for the basic steps when performing IP, or click here for a step-by-step video protocol in JoVE.
High quality affinity reagents are critical for investigatng low abundance PTM modifications
--Utilize PTM affinity reagents that capture a robust profile of the PTM of interest
--PTM reagents that can investigate endogenous changes in the PTM state is beneficial
--PTM IP vs Target Protein IP for enrichment (see below)
--Antibody conjugated to affinity matrices signficantly reduces heavy and light chain leaching
Most investigators work with non-denaturing lysis buffers (e.g. RIPA) . However, there are situations where alternative buffers are required; for example, when trying to isolate DNA bound proteins like histones.
--Choosing the appropriate buffer system may make the difference in a positive or negative PTM identification
--One example, SUMOylation requires investigation in a denaturing buffer system
--Other PTMs are more stable, but may also benefit from denaturing conditions
--Click here to see example data comparing different lysis buffer systems.
PTM inhibitors are chemicals that are supplemented into lysis buffers to inhibit isopeptidases that target specific PTMs (e.g. deubiquitinase inhibitors).
--Identifying key de-PTM inhibitors are critical for successful detection of transient PTMs.
--When investigating multiple PTMs it is important to supplement the lysis buffer with the necessary inhibitors, and it may be necessary to validate any results seperately to ensure no cross reactivity from the cocktail of inhibitors.
--Click here to see example data where cells are lysed with and without the appropriate SUMO isopeptidase (deSUMOylase) inhibitors.
Choose the appropriate control beads:
Control beads allows investigators to distinguish between a real PTM signal, or a false-positive signal from "sticky" proteins that bind to affinity matrices non-specifically.
--Appropriate control beads have IgG antibody bound to the beads to best replicate conditions where any non-specific interactions would be detected.
--Appropriate control beads may also be useful for pre-clearing lysate. Click here to see example data.
Appropriate controls allow the investigator to more easily interpret their results.
--Key controls include positive and negative controls, which are important to interpret results
--Based on information in the literature it may be possible to identify conditions where the target protein of interest is known to be modified or unmodified
--One recomended negative control may be to lyse your cells in buffer without PTM inhibitors, which should result in a dimisnished or absent signal.
--Click here to see a published example of a PTM modified protein where the modification is diminished in the absence of PTM inhibitors.
--Another important control is an HRP labeled pan-antibody against the target PTM. This can be used to evaluate the level of PTM enrichment achieved.
While colorimetric and fluorescent detection methods may provide sensitive, linear western signals for the detection of your target protein, we highly recommend the use of the ultrasensitive chemiluminescence detection reagent as it is generally 10 fold more sensitive than fluorescence detection and 20 fold more sensitive than colorimetric.
High quality primary antibodies are very important.
-- Do not use primary antibodies with high background
-- Do use primary antibodies with high sensitivity
-- Signal validation with multiple antibodies is beneficial
IP is a critical step in the majority of PTM detection techniques; thus, having optimized, high-quality IP reagents will provide the best likelihood of obtaining meaningful results. These reagents include the appropriate lysis buffer system, antibody conjugated to affinity beads (to minimize antibody contamination), control beads, wash buffer, and elution buffer. Cytoskeleton offers the first comprehensive PTM Detection Kits to simplify investigation and detection of thee crucial modifications for any target protein. These kits include the aforementioned, essential tools in order to minimize pitfalls and optimization time, while increasing the rate of novel discoveries.
Protein antibody specific IP: western blot analysis
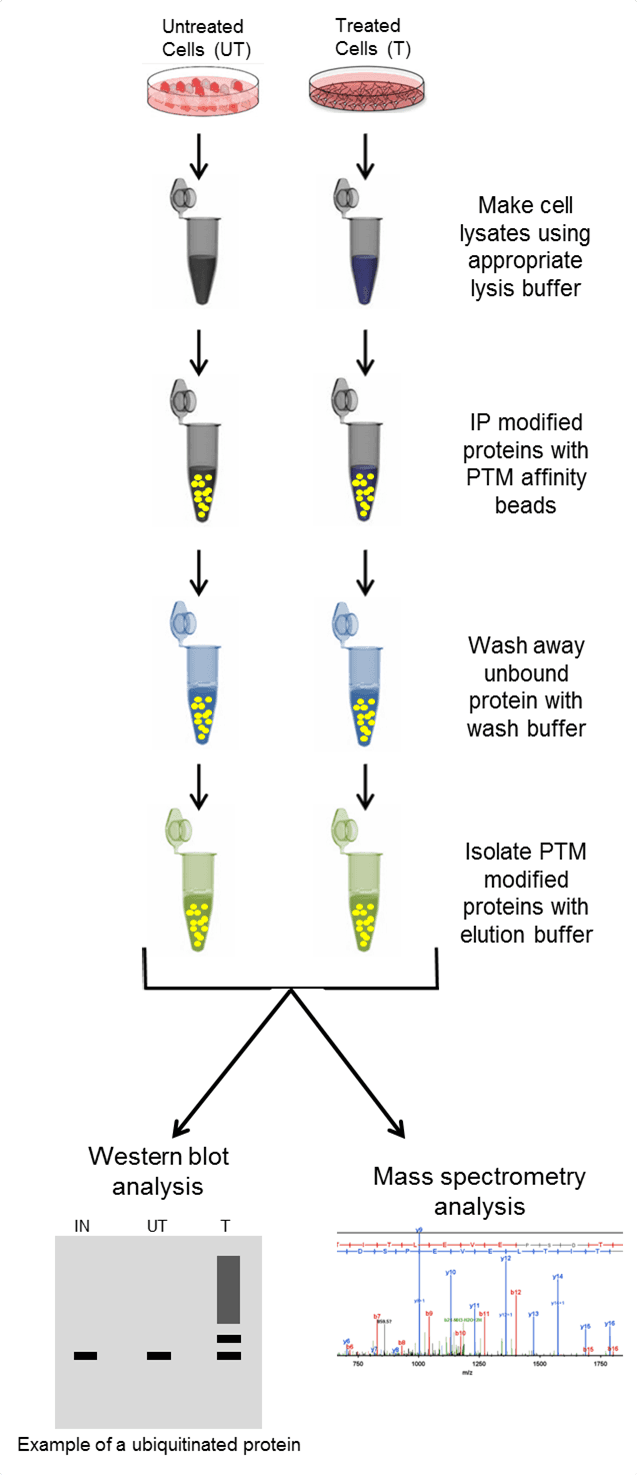
Protein specific IP utilizes an antibody against a POI to immunoprecipitate potentially all species of that protein. The enriched proteins are then separated by SDS-PAGE, transferred to a PVDF membrane, and analyzed via western blot, by probing with a target PTM antibody. Figure 2 provides example data utilizing POI-antibody specific IP, where phosho-SRF was detected using an SRF antibody for IP follwed by phospho serine antibody detection [3]. Target protein antibody IP has traditionally been the first approach used to determine if a POI is modified by a particular PTM, possibly due to the investigator's expertise with the POI specific antibody for western blot applications.
However, as Figure 1 shows, utilizing the antibody in an IP assay is quite different than its use in a western blot application. It is not surprizing that often times IP with a POI specific antibody fails, or requires significant optimization because it is performing a different function in an IP vs western blot application; therefore, as a starting point ensure that the POI specific antibody has been validated for IP. For initial discovery experiments, performing an IP with a PTM specific antibody may be an easier approach, where IP is performed with a PTM specific antibody and the POI specific antibody is used for western blot analysis. This approach only requires optimization of the IP step rather than having to optimize both the IP and the western steps which may be required in a protein specific IP.
Another point of consideration, is that previous research has shown that PTM modifications may block the antibody binding site on a POI; thus, false negatives are possible [4]. Either PTM specific IP or ovexpression IP may circumvent this problem, and should be considered if target antibody IP fails. See below for details on these two approaches.
Pros:
Cons:
PTM antibody specific IP: western blot analysis
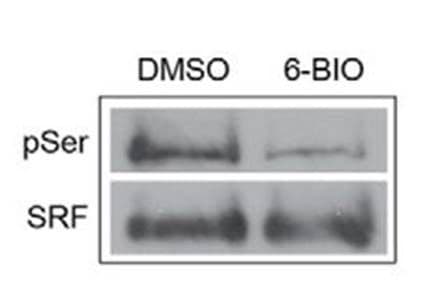
PTM specific IP utilizes an antibody against a target PTM to immunoprecipitate potentially all proteins that have been modified by that PTM (dependent on the quality of the PTM affinity reagent). The enriched post-translationally modified population is then separated by SDS-PAGE analysis, transferred to a PVDF membrane, and analyzed via western blot, by probing with an antibody targeting a specific POI.
Figure 3 shows an example of PTM specific IP using tyrosine phosphorylation, ubiquitination, acetylation, and SUMOylation2/3 PTM affinity beads. The EGFR PTM profile of these four modifications was analyzed by probing with an EGFR antibody. This methodology is great for initial discovery of novel PTMs for any POI. PTM specific IP enrichment is also commonly used for bottom-up proteomics (see below for details), and may be benefical when investigating global PTM changes [5].
The PTM specific IP approach is particularly benfical as kits are available in this format, which allows the investigator to bypass issues that arise with protein specific IP. For example, in a kit format, the antibody is routinely conjugated to beads to minimize heavy and light chain leaching. The Signal-Seeker PTM detection kit in particular provides investigators with key inhibitors, optimized buffer systems, and filters to enhance isolation of modified proteins, and reduce the optimization time. A significant benefit of the Signal-Seeker kits is its ability to capture low-abundance, endogenous PTMs. This feature of the kit was highlighted in a recent publication investigating the clinically relevant PD-L1 protein [6].
Click here for a detailed protocol for PTM specific IP using Signal-Seeker™ PTM detection kits.
Pros:
Cons:
For more information on additional assays please Click here for our PTM detection techniques page.
© 2026 Cytoskeleton, Inc All Rights Reserved.