Abstract
Poly-ubiquitination of a target protein is a well-established mechanism to regulate proteasome mediated degradation; additionally, both poly- and mono-ubiquitination have been shown to control critical protein functions like protein: protein interaction, spatial regulation, and crosstalk with other post-translational modifications (PTMs). Because of its ubiquitous nature, and its clear importance in regulating an array of protein functions, it is no longer practical to leave the investigation of ubiquitination (Ub) to just a few ubiquitin experts. However, user-friendly tools are needed in order for non-Ub-specialists to efficiently study whether and under what conditions, their target protein is ubiquitinated. Here we describe a newly developed Signal-Seeker Ub kit (BK161) that rapidly identifies both poly- and mono-Ub for any target protein. To highlight the utility of this kit, detection of endogenous poly- and mono-Ub of Rac1 as well as chain specific ubiquitin detection was performed. In particular we compare the kits ubiquitin affinity beads, UBA01-beads with other commercially available ubiquitin affinity reagents. Collectively, these studies show that UBA01 is the most robust and precise mono- and poly-Ub detection reagent available, and utilization of the comprehensive BK161 kit will provide the best path to determining if a target protein is ubiquitinated.
Introduction
PTMs like ubiquitin have been shown to regulate many different cellular processes and disease pathologies (2-4).PTM modifications are so abundant that they increase the number of unique protein-forms (or proteoforms) from about 30,000 gene products to over a million proteoforms (5). Surprisingly, the investigation of a target protein PTM profile is not yet viewed as a standard requirement for basic protein and pathway characterization. This is, in part, due to a paucity of user-friendly tools.
Signal-Seeker™ Kits were developed to allow the non-PTM or pro-teomics specialists the ability to gain critical insight into the endogenous form of their protein of interest in less than one day with little or no specialized method development.
Signal-Seeker™ Kit BK161 provides a comprehensive set of reagents to determine the Ub state of any endogenous protein of interest. The kit includes ubiquitin affinity beads UBA01 which are based on a proprietary formulation of Ubiquitin Binding Domain proteins (UBDs). This white paper presents results of a comparison between UBA01 beads and other well established commercially available ubiquitin enrichment reagents.
Results & Discussion
UBA01 is a superior Reagent for the detection of monoubiquitinated proteins
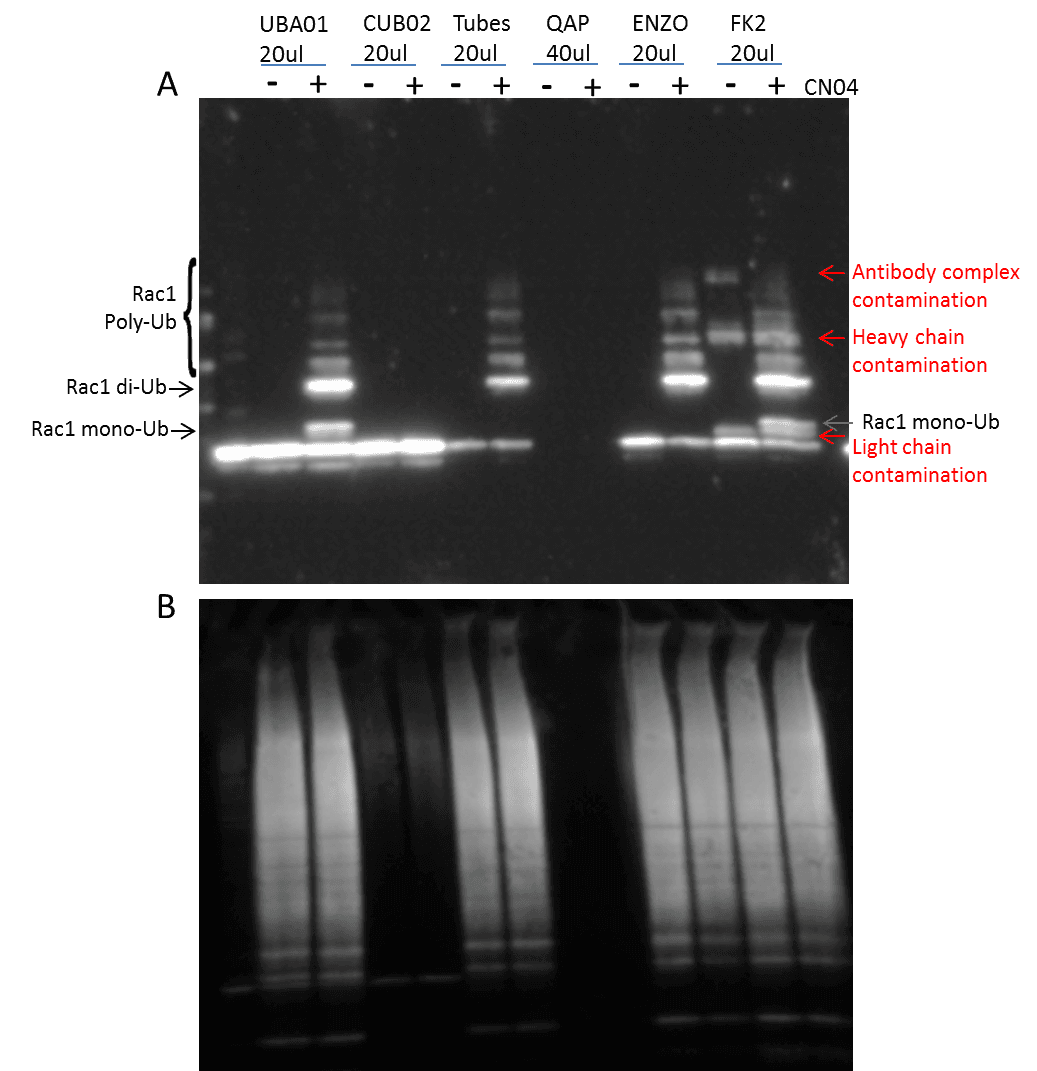
The identification of both poly– and mono-Ub of a target protein is critical, as both types of modification are known to have essential regulatory functions (2). The cytosolic necrotizing factor 1 (CN04) has been shown to induce Rac1 mono– and poly-Ub in the presence of MG-132 (6), and was used as a proof-of-principle, ubiquitin-modified target protein. 3T3 cells were pre-treated with MG-132 followed by CN04 treatment, at which point they were then lysed with BlastR™ buffer (a component of BK161 kit), and immunopre-cipitated with UBA01-beads (Ub affinity beads & a component of (BK161) kit), CUB02-beads (Ub control beads & a component of BK161 kit), Tubes (Lifesensors UM401, agarose-Tube 1), QAP (ENZO, UBIQAP-TURE-Q kit, BML-UW8995-0001), ENZO (ENZO, DSK2 UBA do-main, BML-UW9835-0500), and FK2 (MBL Life science, anti-multi ubiquitin mAb-agarose, D058-8). Samples were then separated on an SDS-PAGE gel and transferred to PVDF. Rac1 Ub was examined using a Rac 1 antibody (Figure 1).
The data shows that UBA01-beads beads and FK2 effectively captures both Rac1 mono– and poly-Ub species which have previously been reported to occur in Rac1 (6). However, the Rac-1 Ub profile captured with UBA01-beads was much easier to interpret compared to the FK2 captured profile, as the UBA01-beads beads did not suffer from the heavy and light chain contamination that commonly occurs with an antibody-based immunoprecipitation system (note Fig 1 red arrows).
Importantly, none of the other ubiquitin domain (UBD)-based affinity beads (Tubes, QAP, ENZO) were able to identify the mono-Ub of Rac1, which is problematic, as mono-Ub is a critical regulatory mechanism for a multitude of target proteins. This data set clearly establishes UBA01-beads beads as a superior reagent for the study of monoubiquitination.
The BK161 kit also provides CUB02-beads control beads. These are identical to the UBA01-beads beads except for two point mutations that abolish Ub binding. This makes them an excellent control bead which are essential when performing Ub detection experiments, because the unmodified form of many target proteins is often captured during the Ub IP step. Figure 1 clearly shows that CUB02-beads identified the non-specific interaction with the unmodified Rac1, but did not interact with Rac1-Ub providing precise results as to which bands correspond to ubiquitinated Rac1.
Total and chain specific ubiquitin recognition by various Ub detection reagents
Total Ub identification by these various Ub detection reagents was determined by re-probing the Rac1 Ub blot with a ubiquitin-HRP specific antibody (AUB01-HRP) . Figure 1B shows similar total Ub recognition by UBA01-beads, Tubes, ENZO, and FK2. While this data suggests that the ability of these reagents are similar, when cou-pled with Figure 1A an alternative interpretation is that the results from total Ub may be misleading, and should only be used for interpretation when the results are drastically different. For example, when comparing UBA01-beads Ub enrichment compared to UBIQAPTURE-QAP Ub enrichment, it is clear that UBA01-beads is capturing a Ub profile where as UBIQAPTURE-QAP is not effectively binding any ubiquitinated species.
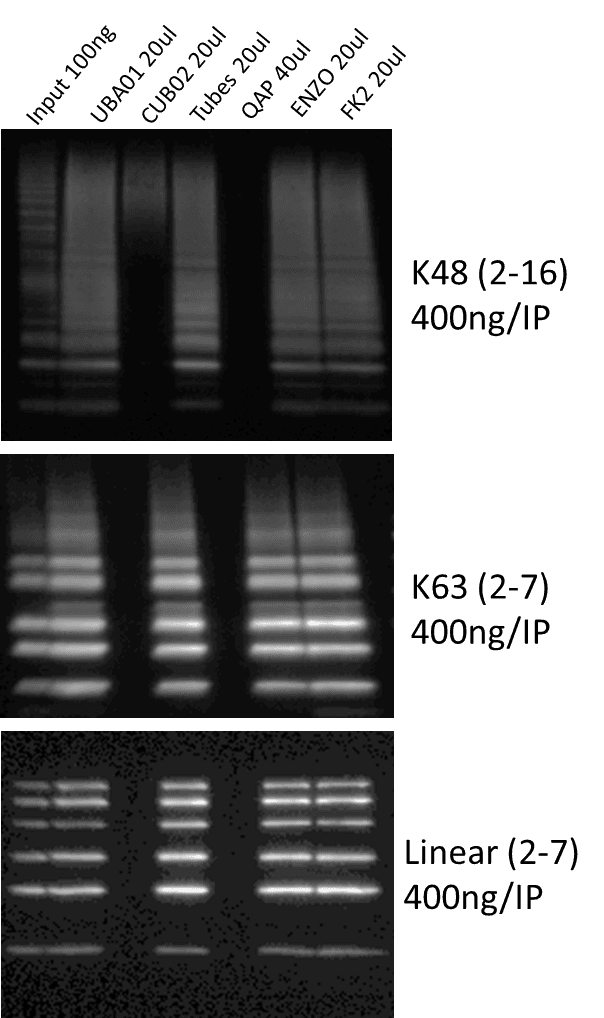
To further investigate the Ub identification capabilities of these reagents, immunoprecipitation (IP) was performed against chain specific ubiquitin. 400ng of K48, K63, and linear ubiquitin was incubated with the various Ub affinity reagents. Samples wereresolved on an SDS-PAGE gel and transferred to PVDF. AUB01 ubiquitin-HRP antibody was used to detect ubiquitin chains (Figure 2). Similar to the total ubiquitination profiles seen in Figure 1A, UBA01-beads, Tubes, ENZO, and FK2 all effectively captured the three types of ubiquitin chains. These data suggest that UBA01-beads is as effective as established ubiquitin affinity reagents at capturing saturating amounts of total and chain-specific ubiquitin profiles.
Comparison of UBA01 and FK2 Ub affinity to chain specific ubiquitin
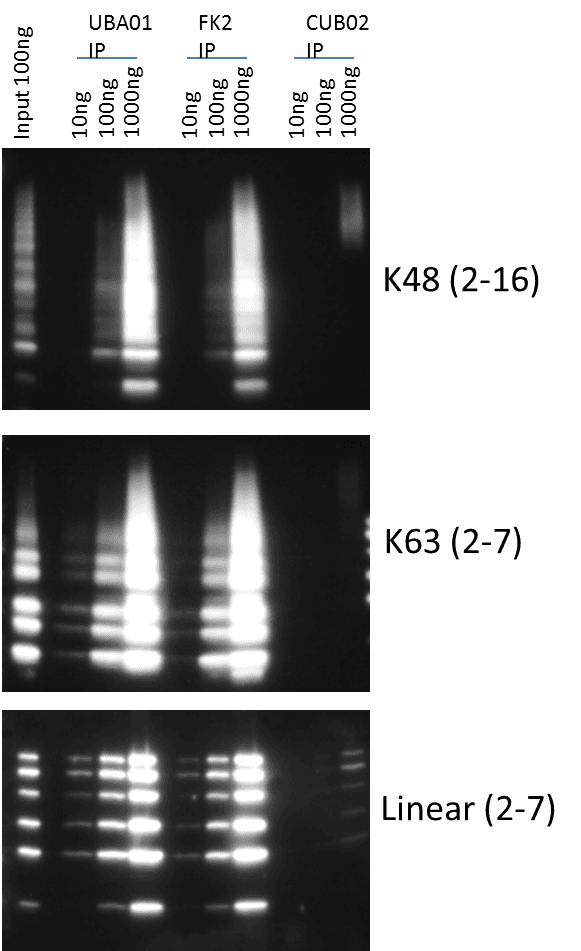
UBA01-beads and FK2 were clearly the most comprehensive ubiquitin affinity reagents as they could capture mono– and polyubiquitinated species including mono-Ub Rac1, and other target proteins (data not shown), further analysis between these two reagents was warranted.
Titrations of k48, k63, and liner ubiquitin chains were used to determine which of these two reagents had a higher affinity for these different ubiquitin chains. Figure 3 shows that UBA01-beads consistently outperformed FK2 for recognition of k48 and k63 ubiquitin chains at low concentrations; thus, providing compelling evidence that UBA01-beads will provide the best method for total Ub detection particularly when detecting low levels of a given ubiquitinated species as is generally the case for endogenous species.
Conclusion
Collectively, these results indicate that the UBA01-beads beads are a superior affinity reagent for the capture of ubiquitinated protein species. The data also shows that the CUB02-beads beads serve as an excel-lent control for non-specific protein binding.
Finally, the UBA01-beads and CUB02-beads reagents have been incorporated into user friendly Signal-Seeker™ Kits which were developed to allow the non-PTM or proteomics specialists the ability to gain critical insight into the endogenous PTM profile of their protein of interest with little or no specialized method development. The end user need only provide a primary antibody to their protein of interest. The kit contains all other necessary reagents, including lysis buffer, inhibitors, affinity reagents and detection reagents to allow an end user to identify ubiquitination of their protein of interest in less than a day.
Materials and Methods
Cell Culture and Reagents
3T3 cells were grown in DMEM media (ATCC, VA) supplemented with 10% FBS (Atlas Biologicals, CO) and penicillin/streptomycin (ThermoFisher, MA). Trypsin/EDTA was obtained from Gibco (ThermoFisher, MA). Unless otherwise noted, chemicals were obtained from Sigma Chemical Co. (Sigma, MO). Cytosolic necrotizing factor 1 (CN04) was obtained from Cytoskeleton, Inc. (Cytoskeleton, CO). For CN04 stimulation experiments 3T3 cells were stepwise serum restricted for 24 hours with serum free DMEM in order to syn-chronize the cells. The cells were then pre-treated with 10 µm MG-132 (Lifesensors, PA) for 2 hours followed by no treatment or CN04 treatment at 15 µg for 3 hours in individual 15cm dishes (Corning, NY) followed by subsequent lysis with BlastR™ lysis buffer (Cytoskeleton, CO).
Western blotting
Treated 3T3 cells were lysed with ice-cold BlastR™ lysis buffer (Cytoskeleton, CO) containing a cocktail of NEM, TSA, Na3VO4, and protease inhibitors (PIC02) (Cytoskeleton, CO). DNA was removed by passing the lysate through the BlastR filter system (Cytoskeleton, CO). After dilution with BlastR™ dilution buffer, protein concentrations were determined with protein reagent, ADV02 (Cytoskeleton, CO), and measured at 600nm OD. Protein lysate samples were separated using Tris-glycine SDS-polyacrylamide gel electrophoresis (ThermoFisher, MA) and transferred to Immobilon- P membranes (Millipore, MA). Membranes were blocked for 1 hr at room temperature in Tris-buffered saline (10 mM Tris-HCl, pH 8.0, 150 mM NaCl) containing 0.05% Tween-20 (TTBS) and 5% milk (Thrive Life, UT), and then incubated with TTBS solution containing Rac1 (BD Biosci-ences, CA) primary antibody for 1 hour at room temperature (RT). Membranes were washed in TTBS 3x10 minutes, prior to secondary Mouse (Jackson ImmunoResearch Laboratories, PA) antibody for 1hr at RT. Bound antibodies were visualized with horseradish peroxidase-coupled secondary antibodies and chemiluminescent reagent (Cytoskeleton, CO) according to the manufacturer’s directions. For total ubiquitin, AUB01 ubiquitin-HRP antibody (Cytoskeleton Inc, CO) was used at 1:4000.
Co-immunoprecipitation assay
3T3 cells were lysed with ice-cold BlastR lysis buffer containing a cocktail of NEM, TSA, Na3VO4, and protease inhibitors (PIC02). DNA was removed by passing the lysate through the BlastR filter system (Cytoskeleton, CO). After dilution with BlastR™ dilution buffer, protein concentrations were determined with ADV02 and measured at 600nm OD. 300 µg of sample lysate was immunoprecipitated, using UBA01-beads (Cytoskeleton Inc, ubiquitination affinity beads), CUB02-beads (Cytoskeleton Inc., ubiquitination control beads), Tubes (Lifesensors UM401, agarose-Tube 1), QAP (ENZO, UBIQAPTURE-Q kit, BML-UW8995-0001), ENZO (ENZO, DSK2 UBA domain, BML-UW9835-0500), and FK2 (MBL Life science, anti-multi ubiquitin mAb-agarose, D058-8. Samples were immunoprecipitated for 1-2 hr at 4°C on an end-over-end tumbler. After incubation, the affinity beads from each sample were pelleted, and washed 3X with BlastR™ wash buffer. Bound proteins were eluted using bead elution buffer (Cytoskeleton, CO) and detected by western immunoblotting.
For chain specific ubiquitin IP, K48, K63, and linear ubiqui-tin chains were obtained from ENZO life sciences (ENZO, NY) and resuspended according to the manufacturer’s instructions. The immunoprecipitations were performed in NP-40 lysis buffer for 1-2 hr at 4°C on an end-over-end tumbler. After incubation, the affinity beads from each sample were pelleted, and washed 3X with NP-40 lysis buffer. Bound proteins were eluted using bead elution buffer and detected by western immunoblotting.
References
- Seo J, Lee KJ. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol. 2004;37(1):35-44.
- Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399-422.
- Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579-86.
- Weathington NM, Mallampalli RK. Emerging therapies targeting the ubiquitin proteasome system in cancer. J Clin Invest. 2014;124(1):6-12.
- Smith LM, Kelleher NL, Consortium for Top Down P. Pro-teoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186-7.
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Ber-toglio J, Gacon G, Mettouchi A, Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21(5):959-65.