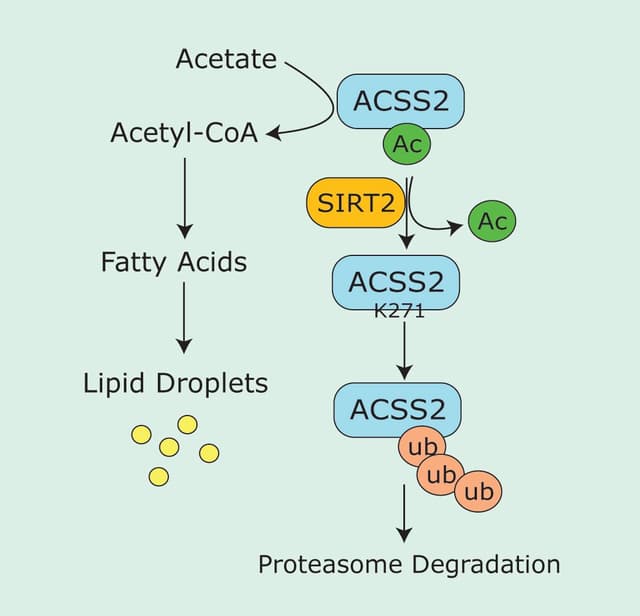
Recent reports have highlighted a new phase of research for molecular motors. In contrast to five or more years ago, when macromolecular approaches were used1, recent research has focused on resolving how kinesin is regulated by intramolecular dynamics. There are two main technical approaches that have allowed measurement of these dynamic mechanisms; these are 1. total internal reflection fluorescence (TIRF) microscopy to observe single kinesin motors and 2. sub-domain swapping from one motor to another.

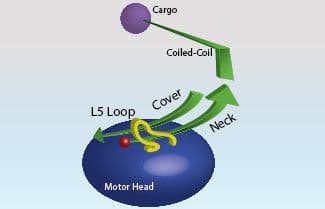
Interestingly, kinesin motor proteins are relatively flexible in accepting substitutions of small amino acid stretches from one partner to another. Kinesins are comprised of two general domains: 1. the catalytic core and 2. the non-catalytic regions which include, the neck linker (NL), coiled-coil (CC) and cargo binding sub-domains. The NL sub-domain is 14-22 amino acids in length (see Figure 1) and Hoeprich et al.2 reported this sub-domain can be swapped from kinesin 1 (K-1) to kinesin 2 (K-2) with measurable function differences being transferred to the new partner. Hoeprich et al.2 used single kinesin tracking to show that the length of the NL sub-domain regulates the ability to move across the microtubule (MT) landscape when MAPs (e.g., tau) are coating its surface. K-2 hardly misses a step when transversing these obstacles, whereas K-1 stutters and progresses more slowly compared to an uncoated MT. In a parameter called processivity or run length, which is a measure of the distance that a motor travels before dissociating from the MT, K-2 travels on average the same distance whether the MT is coated or uncoated with different Tau isoforms (3RS or 4RL), whereas K-1 travels shorter distances on Tau-coated MTs before dissociating. When the NL is genetically swapped between these motors, the characteristic processivity is also swapped.
Similar analyses by Düselder et al.3 highlighted the importance of the NL length in determining processivity of K-5 (also named Eg5 or KIF11). Their results indicate that with a longer NL, e.g., 13-22 amino-acids, processivity is not altered, whereas 11 amino acids and shorter stretches cause decreased processivity. In contrast, velocity and force generation are much less affected by change in length of the NL. However, Shastry and Hancock find dissimilar results with an optimal NL length of 14 amino acids for processivity4,5. The discordant results might be due to different buffers being used, which is noted by Shastry and Hancock as an important determinant of affinity and run length. Interestingly, NL regions have highly specific sequences and lengths for each motor, indicating that this area is optimized for the particular motor’s function. In Table 1, we see that NLs vary from 14 to 22 amino acids and may contain prolines. Prolines are predicted to form a kink in the structure which may cause auto-inhibition of motor activity by bending the coiled-coil back on to the motor domain, or create a different direction of travel for the motor, i.e., enable protofilament hopping or circumferential travel.
Table 1:
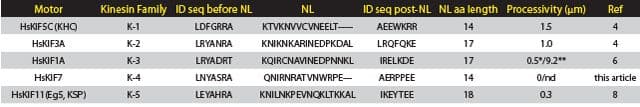
Note: *monomer; **dimer. NL = neck linker; aa = amino acid
Analysis of K-3 family members (e.g., KIF1A, KIF13A, KIF131B, KIF16B) by Soppina et al.6 showed that the coiled-coil 1 sub-domain (CC1) folds back on the NL which caused not only very low processivity but also inhibited dimerization. The typical classification of K-3 members as having low processivity seemed to be contrary to their function as vesicle motors which transport vesicles to distal parts of the cell. Soppina et al.6 showed that replacing the NL-CC1 subdomains with a coiled-coil resulted in a highly processive dimeric construct, hence demonstrating that, in fact, the motors are highly processive when dimerized, which occurs in vivo when they bind cargo7.
Finally, a complex interplay of kinesin sub-domains was reported by Hesse et al.8 K-1 is thought to contain three sub-domains: Cover, NL, and L13 (analogous to L5 in other kinesins)9,10 (Figure 1) which coordinate to generate force from Pi release to the NL, and hence contribute to motor domain movement. By swapping these three sub-domains between K-1 and K-5 family members, Hesse et al.8 discovered some individual features that relate to the specific function of those motors. For example the K-5 members’ domains, when transferred to K-1, conferred the characteristic lower resilience to force representative of K-5 members.
These studies indicate a new level of understanding of kinesin sub-domains, demonstrating that they are interchangeable but also carry specific information about the function of their original motor. These sub-domains are short stretches of amino acids that appear to have a high impact on the motor’s specific functions. These sites are also potential targets for molecular intervention with respect to drug development and molecular biology tools.
Related Products & Resources
Select Products
Myosin - cardiac S1 fragment (Cat. # CS-MYS03)
Myosin - smooth muscle S1 fragment (Cat. # CS-MYS05)
Myosin - skeletal muscle S1 fragment (Cat. # CS-MYS04)
KIF7 motor domain (Cat. # CS-KF51)
Dynein (cytoplasmic) (Cat. # CS-DN01)
Paclitaxel 2mM (Cat. # TXD01)
Kinesin heavy chain motor domain protein (Cat. # KR01)
CENP-E kinesin motor domain protain (Cat. # CP01)
Eg5 kinesin motor domain protein (Cat. # EG01)
Tubulin protein (Pre-formed microtubules) - (Cat. # MT002)
Tubulin protein (Cat. # T240)
Kinesin ATPase Endpoint Assay Biochem Kit™ (Cat. # BK053)
Kinesin ELIPA Biochem Kit™ (Cat. # BK060)
References
- Case R.B. et al. 2000. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 10, 157-160.
- Hoeprich G.J. et al. 2014. Kinesin’s neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 106, 1691-1700.
- Düselder A. et al. 2012. Neck-linker length dependence of processive kinesin-5 motility. J. Mol. Biol. 423, 159-168.
- Shastry S. and Hancock W.O. 2010. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939-943.
- Shastry S. and Hancock W.O. 2011. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253-16258.
- Soppina V. et al. 2014. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. U.S.A. 111, 5562-5567.
- Tomishige M. et al. 2002. Conversion of UNC104/KIF1A kinesin into a processive motor after dimerization. Science. 297, 2263-2276.
- Hesse W.R. et al. 2013. Modular aspects of kinesin force generation machinery. Biophys. J. 104, 1969-1978.
- Khalil A.D. et al. 2008. Kinesin’s cover neck bundle folds forward to generate force. Proc. Natl. Acad. Sci. U.S.A. 105, 19247-19252.
- Hwang W. et al. 2008. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 16, 62-71.