Ultra-Precise HRV C3 Protease (human rhinovirus type 14 3C Protease) (recombinant-GST tagged)
Ultra-Precise HRV C3 Protease (human rhinovirus type 14 3C Protease)
(recombinant-GST tagged)
Background Information
The Ultra-Precise HRV 3C Protease (UP protease) is a fusion protein with a glutathione S-transferase affinity tag (GST) and human rhinovirus (HRV) type 14 3C protease commonly used in molecular biology and protein purification techniques. The UP protease specifically cleaves the specific amino acid sequence Leu-Glu-Val-Leu-Phe-Gln/Gly-Pro (LEVLFQ/GP), cleaving between the Gln and Gly residues. UP protease can maintain robust cleavage activity at 4°C, allowing for improved target protein stability. The GST-tag of UP protease allows for the simultaneous cleavage of a targeted GST-fusion protein substrate and the protease removal when cleaving on Glutathione Sepharose Resin. The cleaved protein substrate can be collected in the flow-through while leaving GST and UP protease bound to the resin. The UP protease is identical in sequence and activity to the popular PreScission protease. The UP protease cleavage site can be found in fusion proteins produced from pGEX-6P vectors.
Material
The recombinant UP protease protein was produced in a bacterial expression system. The recombinant protein has an N-terminal GST-tag and has an approximate molecular weight of 46 kDa. One unit is the amount of enzyme capable of cleaving >90% of 100 µg of fusion protein in 16 hours at 4°C. Protease specific activity is 800-1100 units/mg. The protein is supplied as a white solid.
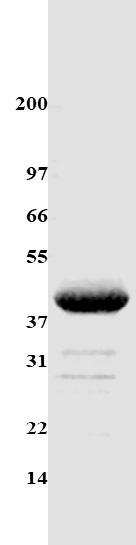
Figure 1. Purity analysis of UP Protease residues Legend: A 10 µg sample of UP protease purity is determined by scanning densitometry of Coomassie Blue-stained protein on a 4-20% polyacrylamide gradient gel. Acetylated tubulin protein was deter-mined to be 90% pure. (see Figure 1). Protein quantitation was performed using the Precision Red Protein Assay Reagent (Cat. # ADV02). Molecular weight markers are from Invitrogen (Mark 12).
Storage and Reconstitution
The recommended storage conditions for the lyophilized material are desiccated at 4°C. Under these conditions, the protein is stable for one year. Lyophilized protein can also be stored desic-cated at -70°C.
Before reconstitution, briefly centrifuge to collect the product at the bottom of the tube. The protein should be reconstituted by adding 100 μl of cleavage buffer for 1 U/µl. When reconstituted, the protein will be in the following buffer: 50 mM Tris pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1.5% sucrose, and 0.2% Dextran. Stable at 4°C for 1-2 weeks.
Cleavage Buffer
The following cleavage buffer needs to be prepared before working with UP Protease.
50 mM Tris-HCl pH 7.0
150 mM NaCl
1 mM EDTA
1 mM DTT
Note: Digestion may be improved by adding 0.01% detergents (Triton X-100, Tween-20, and NP-40). Concentrations up to 1% of detergents do not inhibit protease activities. The presence of Zn2+ has been found to reduce UP protease activity.
Small-scale cleavage optimization of GST-Fusion proteins.
The cleavage activity of UP protease should be optimized on a small scale before scaling up. The amount of UP protease and digestion length for the fusion protein of interest may vary.
Reagents
1. Ultra-Precise HRV C3 Protease (Cat. # CS-UPP01)
2. 100 μg of GST-Fusion Protein
3. Cleavage Buffer at 4°C
Method
1. Resuspend 100U of UP Protease with 100 μl of cleavage buffer.
2. Ensure the GST-Fusion protein has been dialyzed or resuspended in cleavage buffer.
3. Add 100 μg to each of the 4 reaction tubes labeled 2U, 1U, 0.5U, and 0U respectively.
4. Add 2 μl (2 U), 1μl (1U), 0.5 μl (0.5U), and no UP protease as a control to the corresponding reaction tubes. Higher or lower dilution reactions can be tested if desired.
5. Incubate for 16hrs at 4°C.
6. After 16hrs, add 20 μl of reaction to 20 μl of 2X SDS-PAGE loading buffer.
7. Determine cleavage activity by running samples on an SDS-PAGE gel.
8. Reactions can be scaled up after determining ideal digestion conditions.
9. The time for digestion can also be determined with this method with a fixed unit of UP Protease and removing 20 μl samples at different time points.
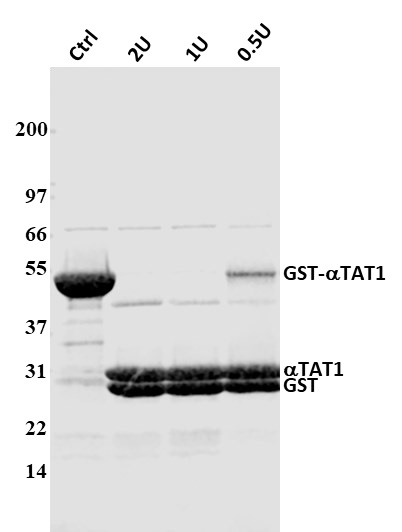
Figure 2. Cleavage of fusion-protein with UP protease Legend: 100 μg of GST-αTAT1 protein was cleaved with UP protease according to the “Small scale cleavage test” conditions in this datasheet. A 20 ug sample of each condition was run on SDS-PAGE, and cleavage efficiency was determined by densitometric analysis of Coomassie blue stained protein bands. Different units of UP were used per reaction (2 units, 1 unit, and 0.5 unit) to determine the amount of enzyme needed for digestion. Molecular weight markers are from Invitrogen (Mark 12).
On-column Cleavage Method
On-resin cleavage of the GST-fusion protein of interest with UP protease is recommended, as the cleaved protein of interest can be separated from GST and UP protease.
1. The recombinant GST-fusion protein of interest can be bound to glutathione resin from cell lysate after the chosen cell lysis method.
2. The glutathione resin can be washed with 5-10 bed volumes of chosen wash buffer to remove protein impurities.
3. Exchange glutathione resin with cleavage buffer and allow the resin to settle. Ensure at least one equal bed volume of cleavage buffer rests above the resin bed. Add 1 to 2U of UP protease per 100 μg of GST-fusion protein and incubate the reaction for 16hrs at 4°C. The reaction can be rotated/shaken slowly overnight to help ensure complete digestion, but specific proteins may precipitate with rotation/shaking versus allowing the reaction to sit overnight with no agitation.
4. The protein of interest can be eluted and collected.
5. The UP protease will remain bound to the glutathione resin.
For product Datasheets and MSDSs please click on the PDF links below.
If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com
Coming soon! If you have any questions concerning this product, please contact our Technical Service department at tservice@cytoskeleton.com
