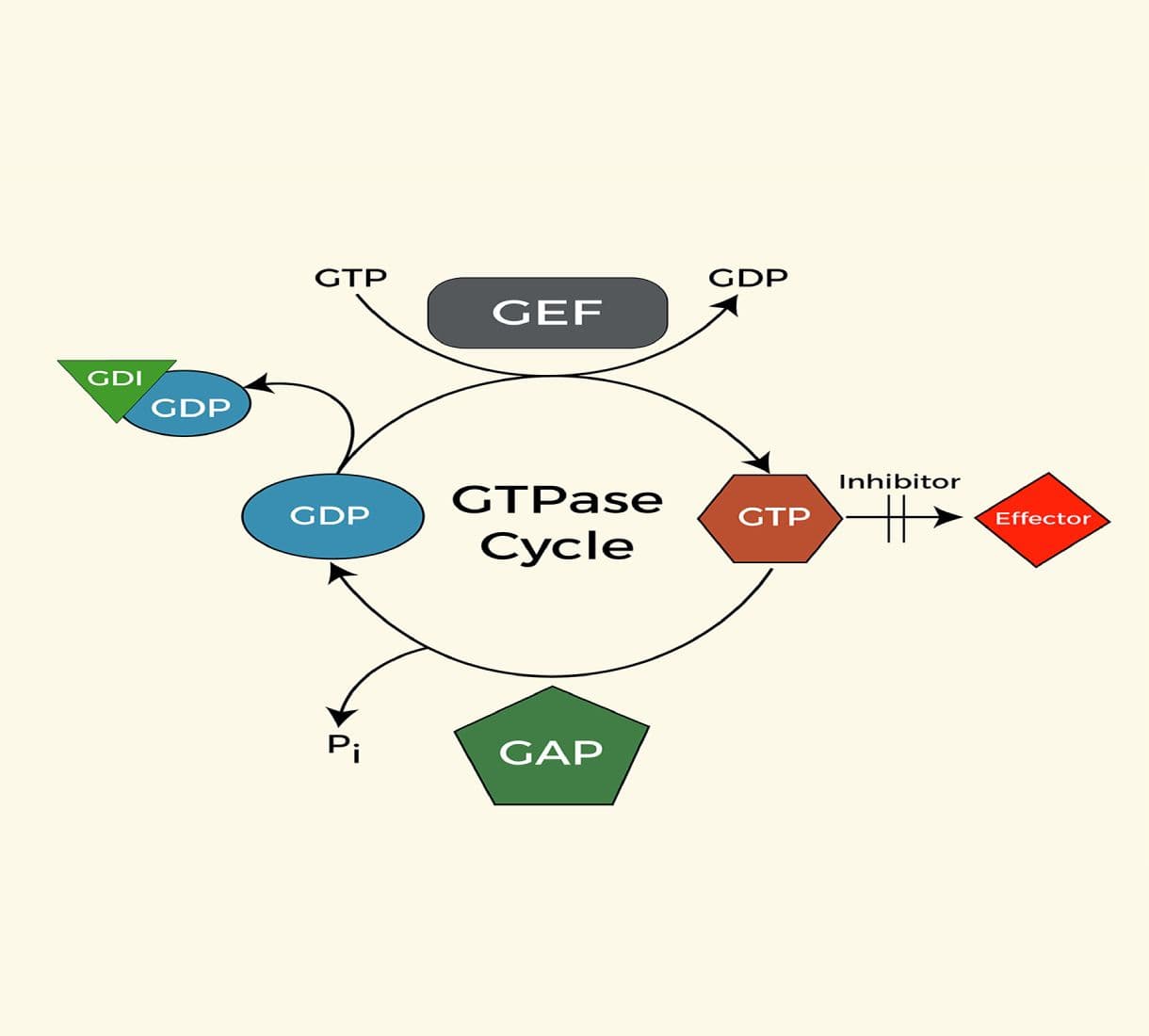
Small G-protein products
Since 1994, Cyto has been at the forefront of small G-protein research.
In consultation with Dr. Alan Hall—a pioneer in the field—we launched the first line of small GTPase reagents. With over 4,000 citations and decades of innovation, we are proud to have contributed to the advancement of small G-protein research.
Today, we continue to support scientists with quality, innovative tools, and expert technical support.