eBook Chapter 5: PTM Functional Characterization and Beyond
Overview of PTMs
Summary of PTMs
Post-translational modifications (PTMs) are dynamic and often reversible alterations to a protein’s structure; importantly, these discrete structural changes have profound, regulatory effects on the protein’s stability, binding partner interactions, and functions (1-3). With the advancement in PTM identification, championed by mass spectrometry (MS) as described in Article 4 of this eBook, and the computational biology field (4-6), scientists now estimate that millions of PTM protein-forms (proteoforms) exist in mammalian cells (7-9).
Clearly PTMs of proteins are prevalent biological processes, but how do we unravel whether a specific proteoform has an essential biological function? Currently, only a very small fraction of proteoforms have been validated via molecular biology approaches and even fewer have been functionally characterized. When identifying key targets from “-omics” data, validation is a critical step, and Article 2 of this eBook highlights effective approaches to validate whether or not a proteoform exists in an investigator’s specific biological model.
Once a promising target has been validated the next step is to determine its functional role in a biological system or disease model. Functional characterization should be performed utilizing a combination of in vitro biochemical assays, molecular biology techniques, and animal-model approaches. Below we describe two examples where biologically-important proteoforms were characterized and highlight essential tools, models, and approaches.
Ubiquitinated Ras Isoforms
The Ras GTPase plays an important role in multiple signal transduction pathways involved in normal cell growth and differentiation as well as several forms of cancer (10, 11). The three isoforms of Ras, H-Ras, N-Ras, and K-Ras, were identified over 30 years ago for their oncogenic activation in human tumors (11). Aberrant Ras signaling has been identified in more than 30% of all human cancers with the most common being lung, colon, and pancreatic cancers (10, 12). In particular, KRas has been identified as the most important Ras protein in cancer research and as such, correlated with over 21% of human cancers (13). Despite extensive research on these proteins, no effective Ras inhibitor has been identified, earning K-Ras the reputation of an undruggable protein.
Ras proteins undergo several post-translational modifications, including proteolytic cleavage by RCE1, farnesylation, carboxymethylation by ICMT, and palmitoylation (14). A seminal study by Jura et al. discovered that HRas protein was mono- and di-ubiquitinated to regulate its localization (15). These studies were done primarily with overexpression of both Ras and ubiquitin proteins in a heterologous CHO-K1 cell line (15). Utilizing these molecular biology tools allowed them to determine that HRas and NRas were ubiquitin substrates while KRas was not. Furthermore, they performed mutagenesis techniques and determined the region of HRas that was ubiquitinated exists in the hyper variable region of the protein, which is distinct from KRas. These studies clearly showed the strength in utilizing molecular overexpression approaches to characterize the ubiquitinated HRas proteoform.
In contrast, a recent study utilizing HEK293T cells determined that KRas can be mono-ubiquitinated, and this modification altered KRas GTP loading and enhanced its affinity for specific downstream effectors (16). The investigators hypothesized that a possible explanation for these opposing findings is that CHO-K1 cells may not have the E3 ligase ubiquitin machinery necessary for K-Ras ubiquitination, and highlights the importance of utilizing multiple biological models during functional characterization. KRas ubiquitination occurred on amino acid K147 as determined by western blotting and mass spectrometry (16). Functional studies on GTP loading and downstream effectors were performed with in vitro biochemical assays and co-immunoprecipitation approaches.
Another interesting result from the aforementioned study in CHO-K1 cells found that HRas di-ubiquitination (di-Ub) was important for HRas activation, but was unchanged in response to upstream EGF regulation (15). In contrast, a more recent study investigating endogenous Ras di-Ub in response to EGF was examined using the Signal-Seeker tools (17). This study reported robust, dynamic changes in di-ubiquitinated Ras in response to EGF stimulation (Figure 1). It is likely that the overexpression system performed in CHO-K1 cells masked physiologic changes in di-Ub Ras; a result postulated by the authors of the original study. Molecular overexpression approaches are indispensable PTM investigation tools as they enable in-depth investigation on site specificity, and isoform specificity as highlighted above; however, we recommend that physiological and biological studies should be validated via endogenous PTM detection approaches. While these studies have conflicting isoform results, they collectively support the original finding that Ras ubiquitination is a biologically significant proteoform; in support of this, a recent study identified the protein LZTR1 as a functional regulator of KRas ubiquitination, and this novel mechanism may contribute to tyrosine kinase inhibitor (TKI) drug resistance (18).

Figure 1: Endogenous ubiquitination of Ras
Figure adapted from Horita et al. 2017. Biosci Rep (17). Serum-restricted A431 cells were stimulated with EGF for the given time period. Unstimulated and EGF-treated A431 lysates were incubated with ubiquitin-affinity beads (UBA01) to immunoprecipitate ubiquitinated proteins or ubiquitin control beads (CUB02). Samples were separated by SDS/PAGE and analyzed by western immunoblotting using a pan Ras antibody to identify ubiquitinated pan Ras. Shown are representative Western blots from n≥3 independent experiments.
Methionine Oxidation of Actin
Actin is a well-characterized, abundant, and essential cytoskeletal protein. Its dynamic properties allow it to shift between monomeric (G-actin) and polymeric (F-actin) states, which is vital for many cellular processes. Actin’s dynamicity and function is regulated by many internal and external cues that are facilitated by actin binding proteins (ABPs), signal transducers, and others. Additionally, several studies now indicate that actin itself is highly modified by post-translational modifications (PTMs). Furthermore, intensive studies of specific actin PTMs have detailed their effect on actin dynamics, ABP interactions, and actin-dependent physiology (19, 20).
A seminal study by the Terman group uncovered a role for the enzyme, MICAL (molecule interacting with CasL), in mediating oxidation of Met44 and Met47 of actin, which they identified with mass spectrometry (21). Functional studies, deduced primarily with elegant in vitro biochemical assays, showed that actin polymerization dynamics was significantly augmented due to oxidation by the redox protein MICAL1. They confirmed these findings with mutagenesis M44L or M47L studies, and found that M44 was essential for MICAL1 regulation of actin polymerization. When oxidized, Met44 becomes negatively charged and interferes with actin monomer-monomer interactions; thus, promoting F-actin severing and depolymerization. Importantly, the group performed studies with an in vivo drosophila model, and found that MICAL1 overexpression had profound consequences, which produced deformed bristle formation that was rescued with M44L actin mutants (21, 22).
The robust results produced in this seminal study has been reproduced by several different groups, and these models and approaches were utilized to identify the counterpart to MICAL1, which is the SelR/MsrB family of methionine sulfoxide reductase enzymes. Two groups independently identified SelR (MsrB) as the enzyme responsible for reduction of Met44 and Met47 and restoration of normal actin dynamics (23, 24). Collectively, the findings identify a reversible and regulatory PTM of actin that controls its dynamics and cytoskeletal organization (Figure 2).
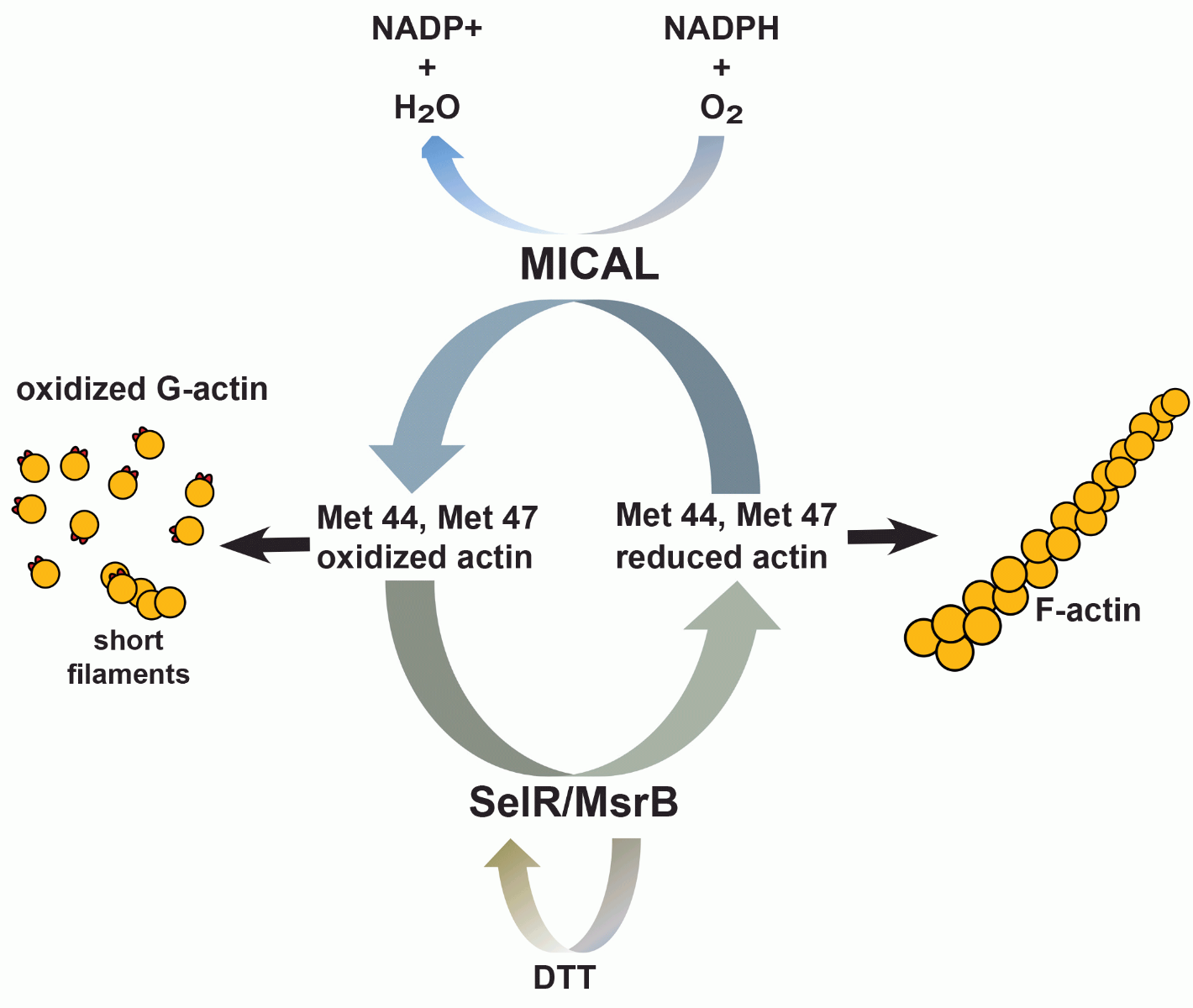
Figure 2: Actin Met44 and Met47 physiological redox system
Since MICALs appear to be expressed ubiquitously and the Met44 and Met47 residues of actin are highly conserved, it is likely that this mechanism of redox regulation may play a prominent role in modulating actin function in all tissue and cell types. A recent study identified MICAL-2 as a regulator of nuclear G-actin levels, subsequent MRTF-A/SRF transcriptional regulation, and physiological regulation of heart development (25). These studies were performed with a combination of in vitro biochemical assays, overexpression models, and in vivo zebrafish studies. Another study investigated the effects that growth factors and chemo-repellents have on MICAL-oxidation regulation of actin, and found paradoxically that effects of the chemo-repellent were amplified by growth factor signaling which had profound effects on physiological axon guidance regulation, as well as pathological tumor progression and response to treatment (26). In totality, these studies identify an actin proteoform that his diverse biological functions.
Conclusions
The examples above identify two critical proteoforms of very distinct proteins, which were found to be important for their respective protein’s function, and when altered, produced profound physiological and pathological consequences. In both cases it took a combination of in vitro biochemical assays, mass spectrometry, overexpression/mutagenesis, endogenous physiologic models, and in vivo models to effectively characterize these proteoforms. The example with Ras ubiquitination highlights potential pitfalls that arise when utilizing a single system with overexpression approaches. While overexpression systems allow investigators to hone in on specific questions regarding form and function of a proteoform, it can mask physiologic mechanisms. Conversely, physiological studies investigating endogenous proteoforms can often provide great correlative data, but gain/loss of function experiments using molecular mutagenesis approaches are useful tools to prove causation and is critical when investigating site specificity for a particular PTM. Ultimately, it is critical to reinforce the idea that utilizing multiple approaches for functional characterization is the best path forward as exemplified with actin oxidation.
Scientist’s understanding of a few PTMs on a select number of target proteins has led to several therapeutic drug targets; still, there are hundreds of thousands of proteoforms waiting to be investigated with in-depth functional studies. The approaches and tools to perform proper biological and physiological validation and characterization are growing, which will simplify the task while providing more accurate and reproducible results that elucidate the importance of these PTMs and proteoforms.
Related Products & Resources
Signal-Seeker™ Ubiquitination Detection Kit (30 assay)
Signal-Seeker™ SUMOylation 2/3 Detection Kit (30 assay)
Signal-Seeker™ SUMOylation 1 Detection Kit (30 assay)
References
- Wu Z, Huang R, Yuan L. Crosstalk of intracellular post-translational modifications in cancer. Arch Biochem Biophys. 2019;676:108138.
- Marcelli S, Corbo M, Iannuzzi F, Negri L, Blandini F, Nistico R, et al. The Involvement of Post-Translational Modifications in Alzheimer's Disease. Curr Alzheimer Res. 2018;15(4):313-35.
- Yan K, Wang K, Li P. The role of post-translational modifications in cardiac hypertrophy. J Cell Mol Med. 2019;23(6):3795-807.
- Gao J, Shao K, Chen X, Li Z, Liu Z, Yu Z, et al. The involvement of post-translational modifications in cardiovascular pathologies: Focus on SUMOylation, neddylation, succinylation, and prenylation. J Mol Cell Cardiol. 2019;138:49-58.
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074-80.
- Liu Y, Tavana O, Gu W. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11(7):564-77.
- Nguyen LK, Kolch W, Kholodenko BN. When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. Cell Commun Signal. 2013;11:52.
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634-7.
- Xie L, Green PL. Envelope is a major viral determinant of the distinct in vitro cellular transformation tropism of human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2. J Virol. 2005;79(23):14536-45.
- Mesnard JM, Barbeau B, Cesaire R, Peloponese JM. Roles of HTLV-1 basic Zip Factor (HBZ) in Viral Chronicity and Leukemic Transformation. Potential New Therapeutic Approaches to Prevent and Treat HTLV-1-Related Diseases. Viruses. 2015;7(12):6490-505.
- Halin M, Douceron E, Clerc I, Journo C, Ko NL, Landry S, et al. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood. 2009;114(12):2427-38.
- Panfil AR, Dissinger NJ, Howard CM, Murphy BM, Landes K, Fernandez SA, et al. Functional Comparison of HBZ and the Related APH-2 Protein Provides Insight into Human T-Cell Leukemia Virus Type 1 Pathogenesis. J Virol. 2016;90(7):3760-72.
- Lowrey AJ, Cramblet W, Bentz GL. Viral manipulation of the cellular sumoylation machinery. Cell Commun Signal. 2017;15(1):27.
- Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11(6):400-11.
- Dubuisson L, Lormieres F, Fochi S, Turpin J, Pasquier A, Douceron E, et al. Stability of HTLV-2 antisense protein is controlled by PML nuclear bodies in a SUMO-dependent manner. Oncogene. 2018;37(21):2806-16.
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450-61.
- Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51(3):221-8.
- Li Y, Li F, Jiang F, Lv X, Zhang R, Lu A, et al. A Mini-Review for Cancer Immunotherapy: Molecular Understanding of PD-1/PD-L1 Pathway & Translational Blockade of Immune Checkpoints. Int J Mol Sci. 2016;17(7).
- Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res. 2013;19(5):1021-34.
- Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409-16.
- Horita H, Law A, Hong S, Middleton K. Identifying Regulatory Posttranslational Modifications of PD-L1: A Focus on Monoubiquitinaton. Neoplasia. 2017;19(4):346-53.
- Horita H, Law A, Hong S, Middleton K. A simple toolset to identify endogenous post-translational modifications for a target protein: a snapshot of the EGFR signaling pathway. Biosci Rep. 2017.

