+3
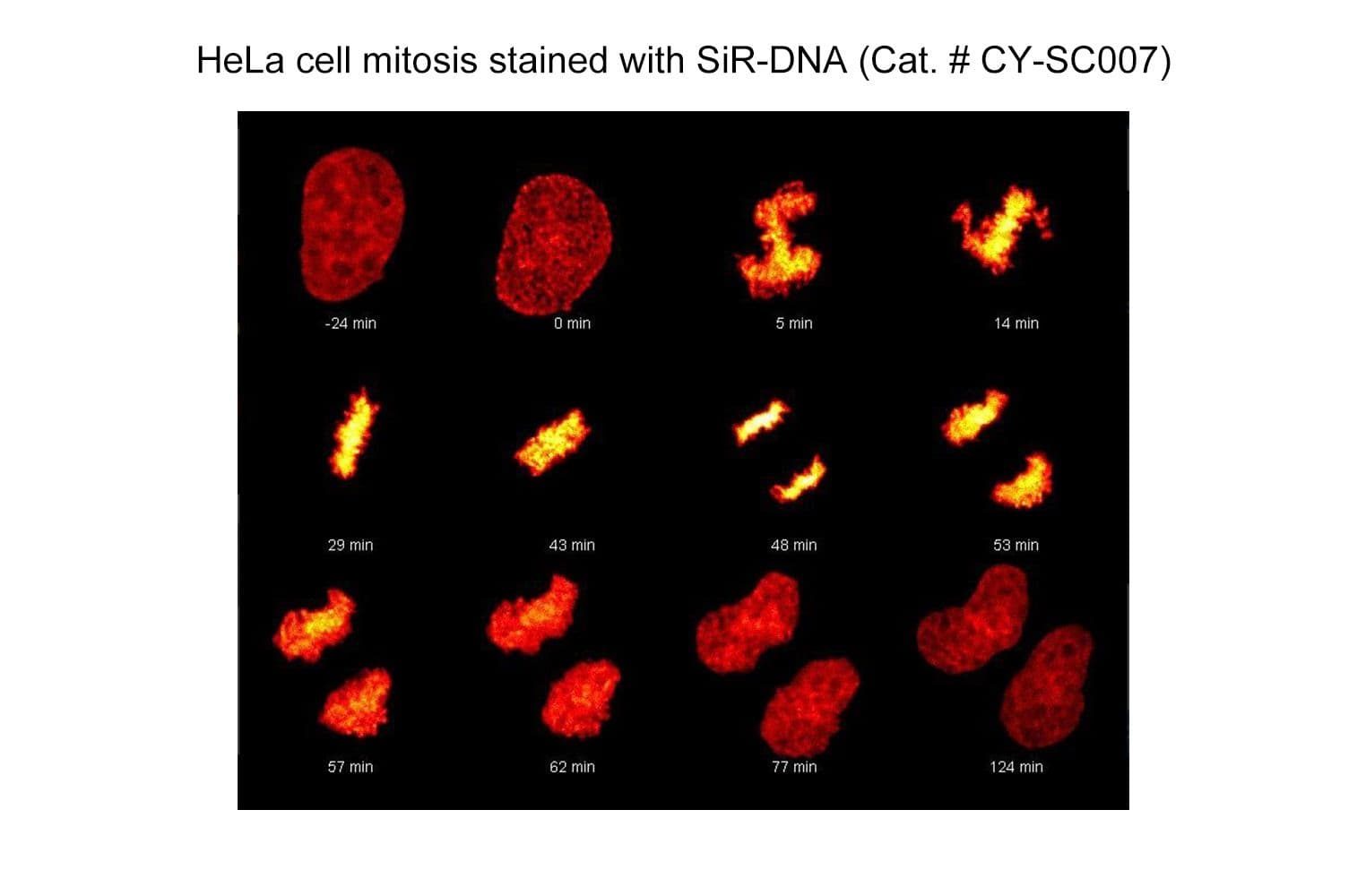
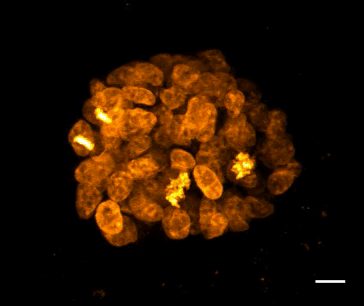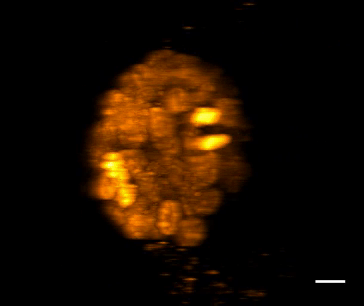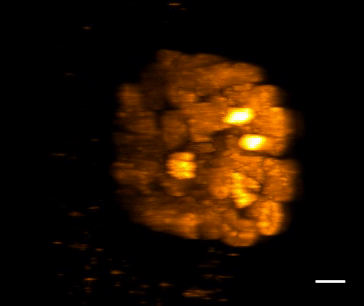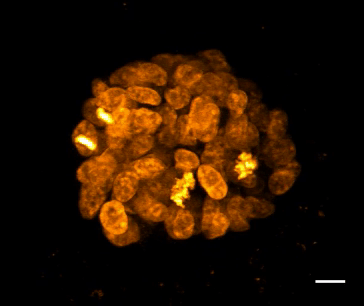
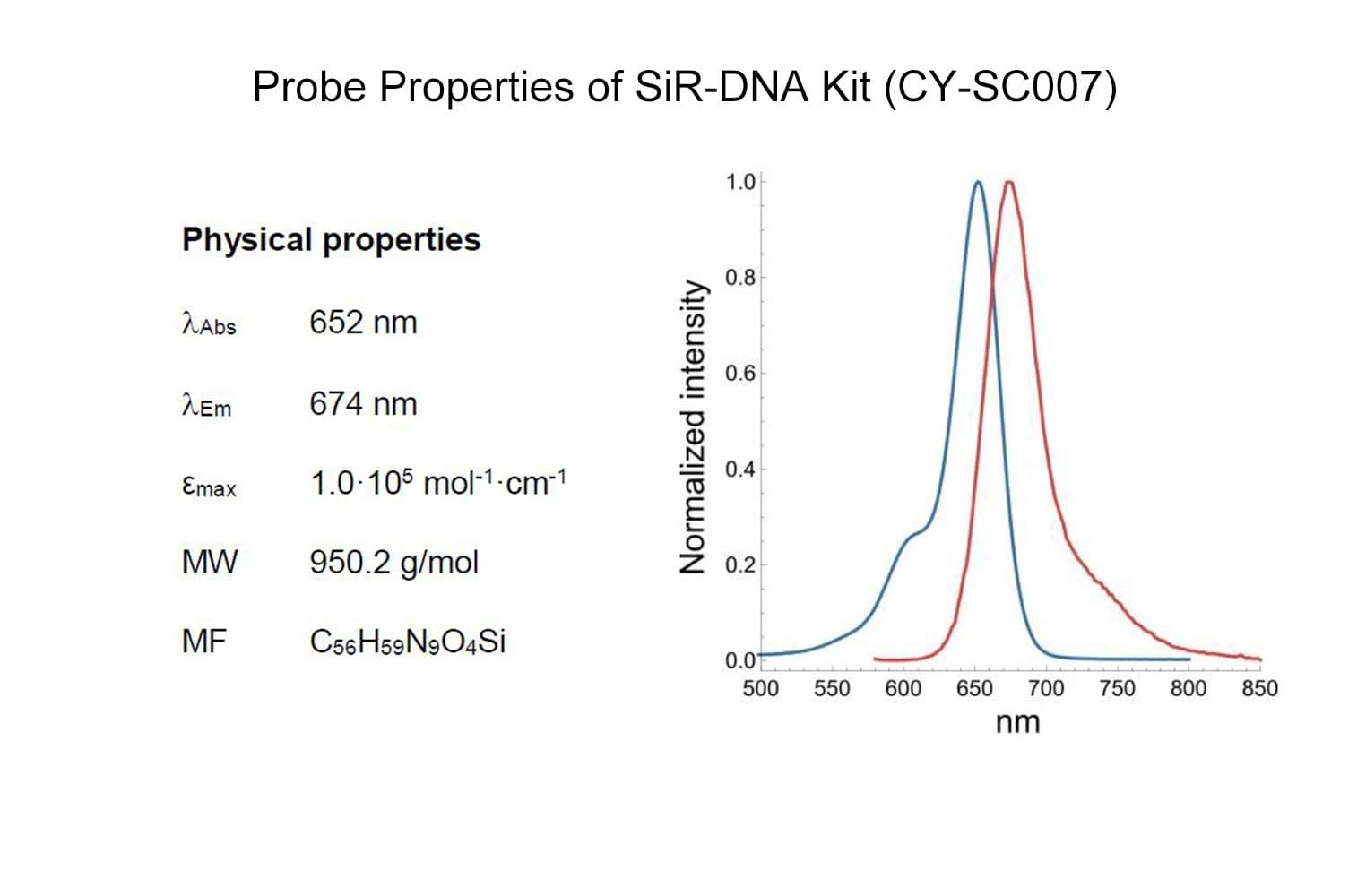
SiR-DNA is based on the fluorophore silicon rhodamine (SiR) and the DNA minor groove binder bisbenzimide (Hoechst). Sir-DNA allows the labelling of DNA in live cells with high specificity and low background.
Key features
The biological activity of CY-SC007 is assessed by the ability of the probe to efficiently label DNA in live human fibroblast cell culture. After an optional wash step, cell staining is visible for several hours. Omit the wash step for time lapse imaging.
Cat. #CY-SC007

© 2026 Cytoskeleton, Inc All Rights Reserved.