+3
Kit contents (30 – 100 assays)
Equipment & materials required
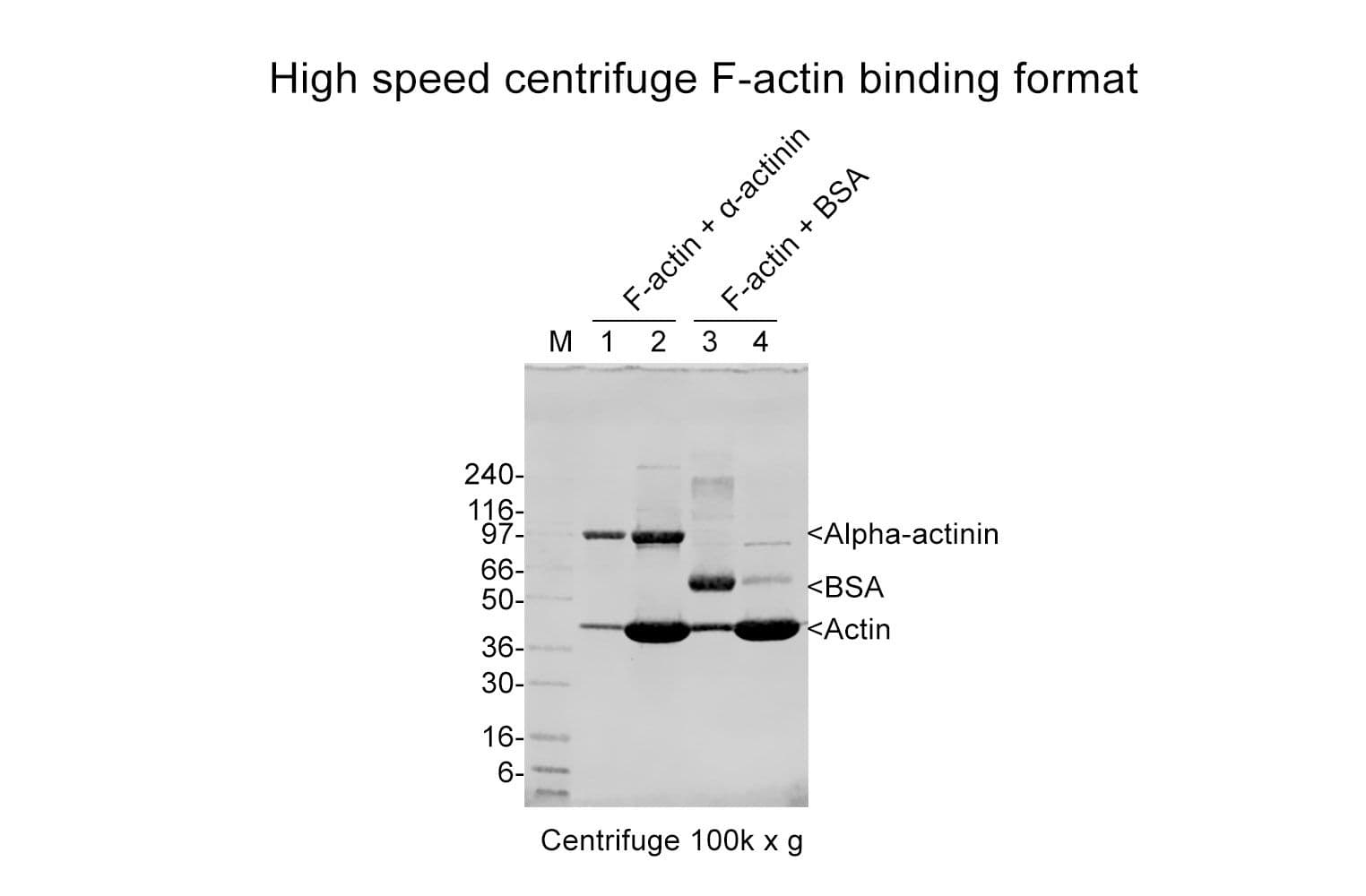
The actin-binding protein (ABP) spin-down assay kit is a convenient and reliable tool for determining whether your protein of interest (POI) binds to filamentous actin (F-actin) or monomeric actin (G-actin). This assay uses a simple centrifugation-based method to separate F-actin and G-actin, allowing you to detect interactions with your POI from either purified protein samples or cell lysates.
Key characteristics
Cat. #BK013