Post-translational Modifications Modulate p53 Tumor Suppressor Functions
The multi-domain, tetrameric p53 protein was discovered in 19791-3, first mistakenly described as an oncogene, before its true function as a powerful tumor suppressor was realized4,5. p53 consists of two N-terminal transactivation domains (TAD1 and TAD2), a proline-rich domain (P-rich), a sequence-specific DNA binding domain (DBD), a linker domain, tetramerization domain (TD), and a lysine-rich, basic C-terminal regulatory (REG) domain5 (Fig. 1). As a transcription factor, p53 can regulate the expression of up to 3000 genes involved in apoptosis, senescence, cell cycle arrest, DNA repair, apoptosis, tumor microenvironment, autophagy, and invasion/metastasis6-8. p53 functionality is spatiotemporally regulated by up to fifty post-translational modifications (PTMs)that occur within multiple domains9-12 (Fig. 1). Here, regulation of p53 by ubiquitination, phosphorylation, and acetylation is discussed.
Ubiquitination
During normal cell growth and development (i.e., non-stress conditions), the MDM2 (mouse double minute 2 protein, mouse homolog of human protein HDM2) E3 ligase maintains p53 expression at a minimal level by ubiquitin-proteasome-regulated degradation through a protein complex consisting of MDM2, p53, and p300/CREB binding protein (p300/CBP)13,14. MDM2 binds p53's TAD1 and negatively regulates p53 as it mediates poly-ubiquitination on at least six different lysine residues within p53’s REG domain15 (Fig. 1). Importantly, MDM2 directly mediates mono-ubiquitination at multiple sites on p5316, but is not directly responsible for poly-ubiquitination. Instead, p300/CBP, which has both ubiquitin ligase and acetyltransferase activity, subsequently utilizes these mono-ubiquitinated lysines as substrates for poly-ubiquitination13,14. Notably, numerous other E3 ligases, such as Pirh2, COP1, CHIP, ARF-BP1, E6-AP, TOPORS, TRIM24, and MKRN1, are also capable of regulating p53 turnover9.
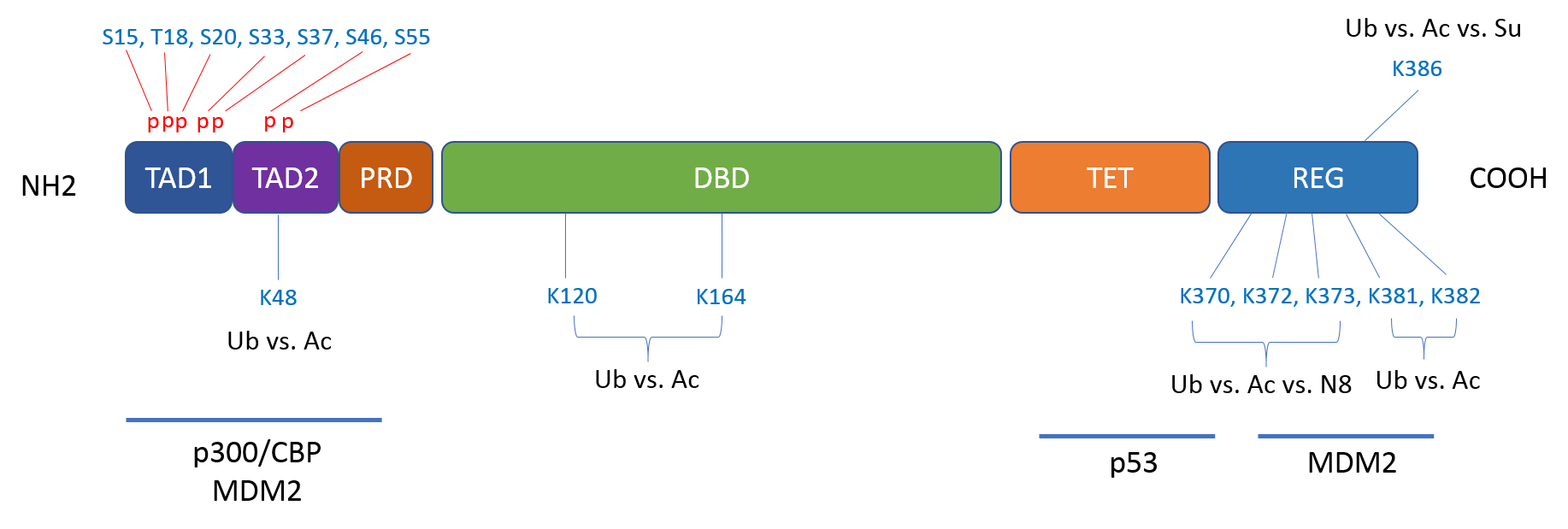
Figure 1. PTMs of p53. p53 is a substrate of approximately 50 PTMs within multiple domains. The PTMs show interdependence, cooperativeness, and/or mutual inhibition.
Phosphorylation
Phosphorylation of serine/threonine (Ser/Thr) residues stabilizes and activates p53 and is triggered by cellular stresses, including genotoxic damage, oncogene activation, hyperproliferative signals, hypoxia, and nutrient starvation17,18. When activated, tetrameric p53 forms active transcriptional complexes on target gene promoters to regulate expression of genes involved in cell cycle arrest and apoptosis. Arguably, the most important phosphoepitope governing p53 activation is Ser15 as its phosphorylation is required for subsequent phosphorylation of other p53 residues (e.g., Thr18 and Ser20)9,19. In fact, Thr18 phosphorylation is the critical catalyst for disruption of MDM2 binding. Phosphorylation of neighboring Ser residues (Ser20, -33, -37, -46, and -55) within the TAD1 further reduces MDM2 binding affinity to p539,19-26 (Fig. 1). Concomitant with this loss of MDM2 binding is the recruitment and binding of the co-transcriptional activator p300/CBP9,19, whose binding with p53 is also critically reliant upon Ser15 and Thr18 phosphorylation21,22. p300/CBP has four structurally similar domains (TAZ1, KIX, TAZ2, and NCBD) that bind p53’s TADs with varying degrees of affinity24-26. Upon di-phosphorylation of p53, bound MDM2 dissociates and p53’s affinity for the KIX domain is mildly enhanced, and affinity for both KIX and TAZ1 domains is significantly increased by sequential phosphorylation of the neighboring Ser residues (~80 fold, compared to Thr18)9,24-26. Indeed, p300/CBP binding to, and activation of, p53 occurs along a gradient which increases with multi-site phosphorylation of p5322,24,26.
Acetylation
Besides phosphorylation, p300/CBP-mediated acetylation is essential for p53 stabilization and activation in response to cellular stresses27-29. At least six lysine residues (Lys370, Lys372, Lys373, Lys381, Lys382, and Lys386) within the REG domain of p53 are targeted for acetylation by p300/CBP. This prevents MDM2-mediated ubiquitination of these lysines and maintains p53 transactivation capacity9,28. Also, p53 acetylation is indispensable for destabilizing the p53-MDM2 interaction, regardless of phosphorylation status. Acetylation of two lysines (Lys120 and Lys164) in p53’s DBD and six lysines in the REG domain blocks the recruitment of MDM2 to the p53 complex binding promotor and enables the p53-mediated stress response29 (Fig. 1). MDM2 promotes p53 deacetylation through the recruitment of a HDAC1 complex28, which allows for poly-ubiquitination of p53 and eventual p53 degradation by the proteasome.
Summary
The tetrameric p53 tumor suppressor protein regulates the expression of thousands of genes involved in cell cycle arrest, apoptosis, and associated pathways. Thus, it is tightly negatively regulated by MDM2- and p300/CBP-mediated poly-ubiquitination. Cellular stresses disinhibit p53 through phosphorylation- and acetylation-mediated stabilization and activation. HDAC1-mediated deacetylation returns p53 to a quiescent state until the arrival of the next cellular stress. To better understand how these and other PTMs can regulate p53 and other cancer-related proteins, Cytoskeleton, Inc. offers Signal-Seeker Enrichment Kits to study the endogenous levels of tyrosine phosphorylation, ubiquitination, acetylation, and SUMOylation in cell and tissue lysates, as well as PTM antibodies validated for multiple applications.
Related Products & Resources
Signal Seeker™ Kits
New Signal-Seeker™ Acetyl-Lysine Detection Kit (30 assay) (Cat. # BK163)
New Signal-Seeker™ Acetyl-Lysine Detection Kit (10 assay) (Cat. # BK163-S)
Signal-Seeker™ Phosphotyrosine Detection Kit (30 assay) (Cat. # BK160)
Signal-Seeker™ Phosphotyrosine Detection Kit (10 assay) (Cat. # BK160-S)
Signal-Seeker™ Ubiquitination Detection Kit (30 assay) (Cat. # BK161)
Signal-Seeker™ Ubiquitination Detection Kit (10 assay) (Cat. # BK161-S)
Signal-Seeker™ SUMOylation 2/3 Detection Kit (30 assay) (Cat. # BK162)
Signal-Seeker™ SUMOylation 2/3 Detection Kit (10 assay) (Cat. # BK162-S)
PTM Antibodies, Beads, Etc
New Acetyl-Lysine Antibody Mouse Monoclonal (7B5A1) (Cat. # AAC02)
New Acetyl-Lysine-HRP Antibody Mouse Monoclonal (19C4B2.1) (Cat. # AAC03-HRP)
Phosphotyrosine Affinity Beads (Cat. # APY03-beads)
Phosphotyrosine-HRP Mouse Monoclonal Antibody 27B10 (Cat. # APY03-HRP)
SUMOylation 2/3 Affinity Beads (Cat. # ASM24-beads)
Ubiquitin Antibody Mouse Monoclonal (Cat. # AUB01)
References
- Kress M. et al. 1979. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 31, 472-483.
- Lane D.P. and Crawford L.V. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature. 278, 261-263.
- Linzer D.I. and Levine A.J. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 17, 43-52.
- Soussi T. 2010. The history of p53. A perfect example of the drawbacks of scientific paradigms. EMBO Rep. 11, 822-826.
- Vousden K.H. and Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell. 137, 413-431.
- Bieging K.T. et al. 2014. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 14, 359-370.
- Fischer M. et al. 2017. Census and evaluation of p53 target genes. Oncogene. 36, 3943–3956.
- Kruiswijk F. et al. 2015. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393-405.
- Meek D.W. and Anderson C.W. 2009. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950.
- Kruse J.P. and Gu W. 2009. Modes of p53 regulation. Cell. 137, 609-622.
- Carter S. and Vousden K.H. 2009. Modifications of p53: competing for the lysines. Curr. Opin. Genet. Dev. 19, 18-24.
- Bode A.M. and Dong Z. 2004. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 4, 793-805.
- Grossman S.R. et al. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell. 2, 405-415.
- Grossman S.R. et al. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 300, 342-344.
- Rodriguez M.S. et al. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell Biol. 20, 8458-8467.
- Lai Z. et al. 2001. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 276, 31357-31367.
- Giaccia A.J. and Kastan M.B. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12, 2973-2983.
- Hu W. et al. 2012. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer. 3, 199-208.
- Saito S. et al. 2003. Phophorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 278, 37536-37544.
- Shieh S.-Y. et al. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 91, 325-334.
- Lambert P.F. et al. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048-33053.
- Lee C.W. et al. 2010. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multi-site phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 107, 19290-19295.
- Dumaz N. and Meek D.W. 1999. Serine 15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18, 7002-7010.
- Teufel D.P. et al. 2007. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl. Acad. Sci. U.S.A. 104, 7009-7014.
- Ferreon J.C. et al. 2009. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc. Natl. Acad. Sci. U.S.A. 106, 6591-6596.
- Teufel D.P. et al. 2009. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene. 28, 2112-2118.
- Gu W. and Roeder R.G. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 90, 595-606.
- Ito A. et al. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236-6245.
- Tang Y. et al. 2008. Acetylation is indispensable for p53 activation. Cell. 133, 612-626.

