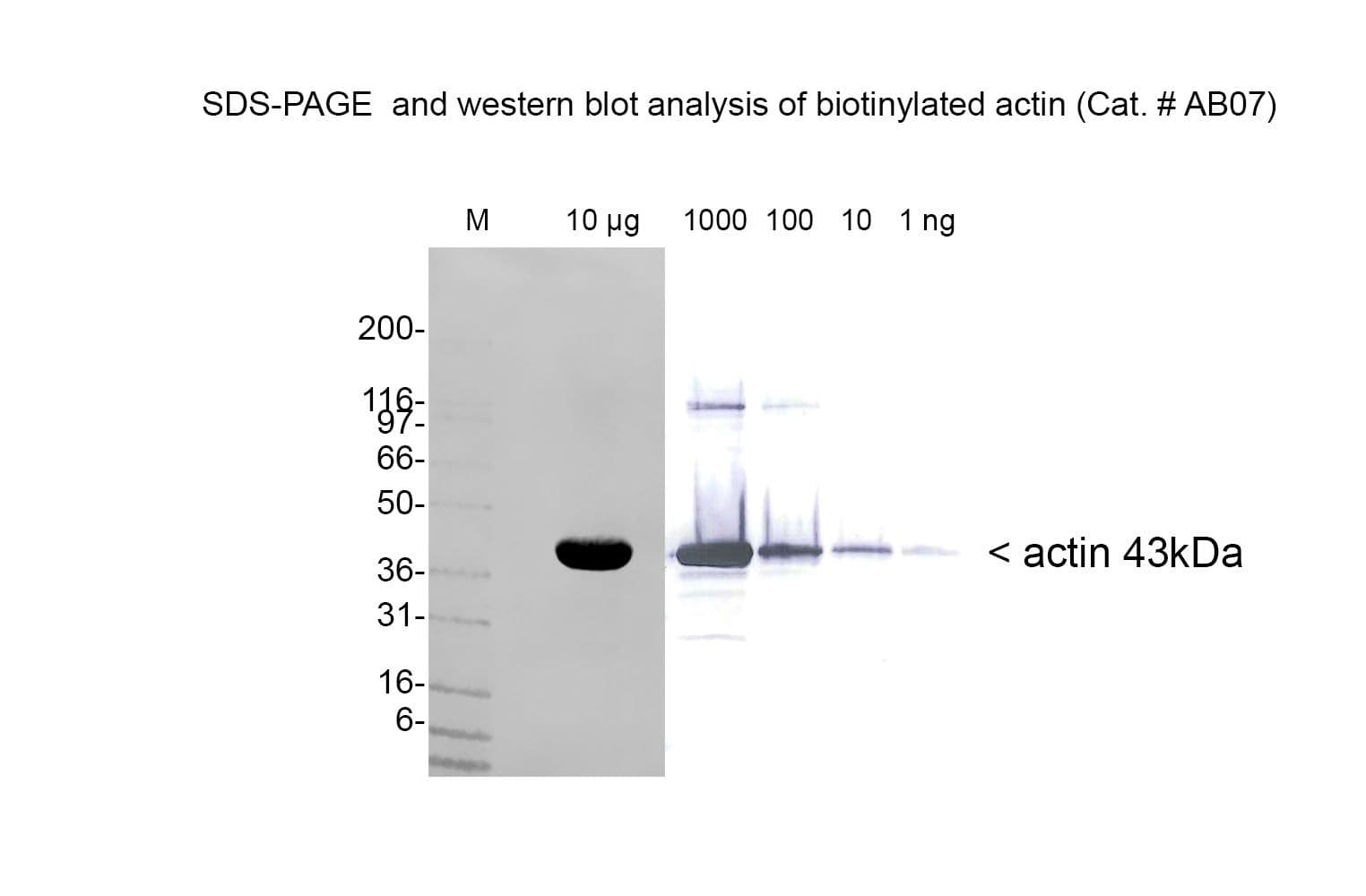
Rabbit skeletal muscle actin AKL99 is biotinylated at random surface lysines using an activated biotin ester. Labeling stoichiometry is ~1 biotin per 43 kDa monomer. It is supplied as a lyophilized powder.
Protein purity is determined by scanning densitometry of Coomassie Blue-stained protein on a 12% polyacrylamide gel. Biotinylated actin was found to be >99% pure.
The biological activity of biotinylated actin has been determined by its ability to polymerize into filaments in vitro efficiently and separate from unpolymerized components in a spin-down assay. Stringent quality control ensures that >90% of the biotinylated actin can be polymerized in this assay. This is comparable to the polymerization capacity of unmodified actin AKL99.
Cat. #AB07