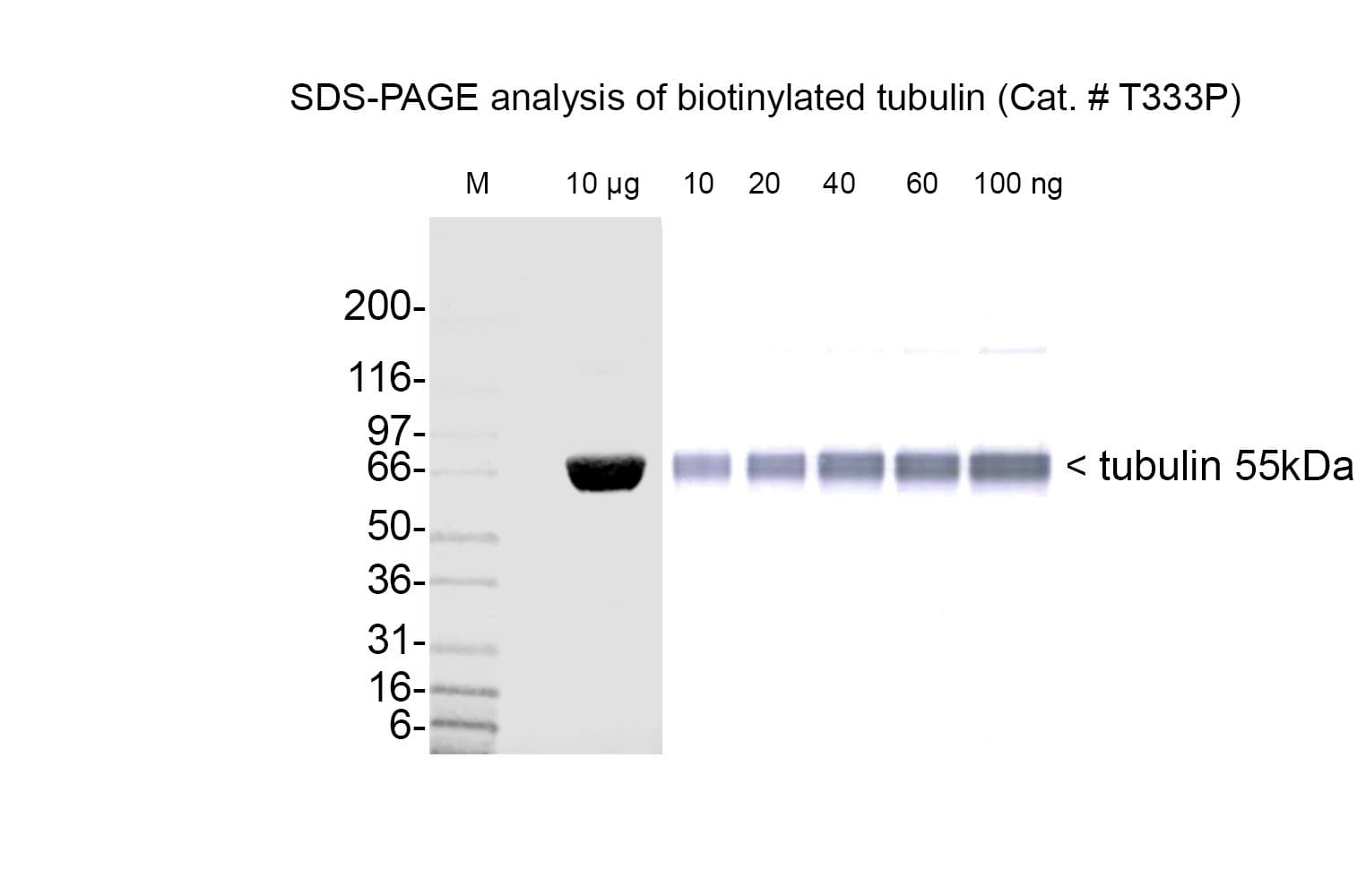
+3
Tubulin protein T240 is labeled on random surface lysines using a long-chain biotin ester. The extended spacer ensures the biotin does not interfere with downstream applications such as streptavidin binding. Labeling yields ~1–2 biotins per tubulin heterodimer. Supplied as a white lyophilized powder.
Protein purity is assessed by scanning densitometry of Coomassie Blue stained protein on a12% polyacrylamide gel. Purity is determined to be >99% pure.
Biotinylated tubulin retains high biological activity, with >80% polymerizing into microtubules in vitro, as confirmed by a spin-down assay. This performance is comparable to that of unmodified tubulin.
Cat. #T333P