Cdc42 is a 21.3 kDa, small GTPase protein encoded by 191 amino acids that belongs to the Rho sub-family of Ras super-family GTPases. There are two isoforms produced by alternative splicing with isoform 1 being the most commonly studied and expressed ubiquitously. Conversely, isoform 2 is restricted to the brain (also known as brain isoform, Cdc42b or G25K), but has been chosen as the 'canonical' sequence. Cdc42 switches between an active GTP-bound state and inactive GDP-bound state. This cycle is regulated by its intrinsic GTPase activity and its interaction with guanine exchange factors (GEFs) which promote the exchange of GDP for GTP and GTPase activating proteins (GAPs) which activate Cdc42’s GTP hydrolysis activity. Cdc42 localizes to membranes via a post-translational C-terminal geranylgeranyl lipid modification; however, it can be found as a soluble complex association with RhoGDI (Rho GDP-dissociation inhibitor), another means of regulating Cdc42’s activation. RhoGDIs sequester Cdc42-GDP in the cytoplasm through a transfer of geranylgeranyl moiety from membrane to GDI and inhibit its spontaneous GDP/GTP exchange activity. Additionally, Cdc42 can be phosphorylated by diverse kinases which regulate its interaction with RhoGDIs. To date over 70 Rho GEFs, 60 Rho GAPs and 3 RhoGDIs have been identified in mammals, reflecting the complexity of regulation of these classes of proteins.
Activation of Cdc42 is part of a signaling cascade initiated by stimulation of cell surface receptors and cell adhesion molecules which include tyrosine kinase receptors, heterodimeric G-protein-coupled receptors (GPCRs), cytokine receptors, integrins, and physical and chemical stresses. After GEF-mediated nucleotide exchange, GTP-bound Cdc42 can interact with a variety of downstream effector proteins, of which at least 23 have been identified. Putative effectors for Cdc42 include: p70 S6 kinase, MLK2 and MLK3, MEKK1 and MEKK4, PAK1, PAK2, PAK3 and PAK4, MRCKα and MRCKβ, ACK1 and ACK2, PI3K, PLD, PLC-β2, WASP and N-WASP, MSE55 and BORGs, IQGAP1 and IQGAP2, and CIP-4.
Cdc42 plays a significant role in a wide variety of cellular processes that are dependent on the actin cytoskeleton, such as cytokinesis, phagocytosis, cell migration, morphogenesis, chemotaxis, axon guidance, axon myelination, intracellular trafficking, gene transcription, cell-cycle regulation and cell fate determination. In regards to cell motility, Cdc42 is directly involved in the formation of filopodia via interactions with actin binding proteins which are nucleation factors). However, the contribution of Cdc42 in these different cellular processes could be cell-type specific. Dysregulation of Cdc42 is found in several pathogenic processes such as cancer, neurodegenerative disorders, and cardiovascular disease.
CCdc42’s role in cancer is dependent on over-expression and/or over-activation rather than activating mutations. Cdc42 contributes to cancer development through defects or dysfunction in its physiological roles. For instance, alterations in Cdc42-mediated intracellular trafficking - especially through the regulation of the processing and degradation of EGF receptor, cell cycle regulation and survival, polarity, migration, and transcriptional control is related to oncogenesis. The contribution of Cdc42 to cancer progression seems to be tissue-specific since Cdc42 appears to have pro-oncogenic and anti-oncogenic properties depending on cellular context. However, there are more cell types where Cdc42 is a pro-oncogenic factor. Cdc42 protein levels are significantly elevated in breast cancer tissue vs normal tissue, testicular tumor tissue, head and neck squamous cell carcinomas, melanomas, colorectal cancer, and hepatocellular carcinoma.
Image of CDC42
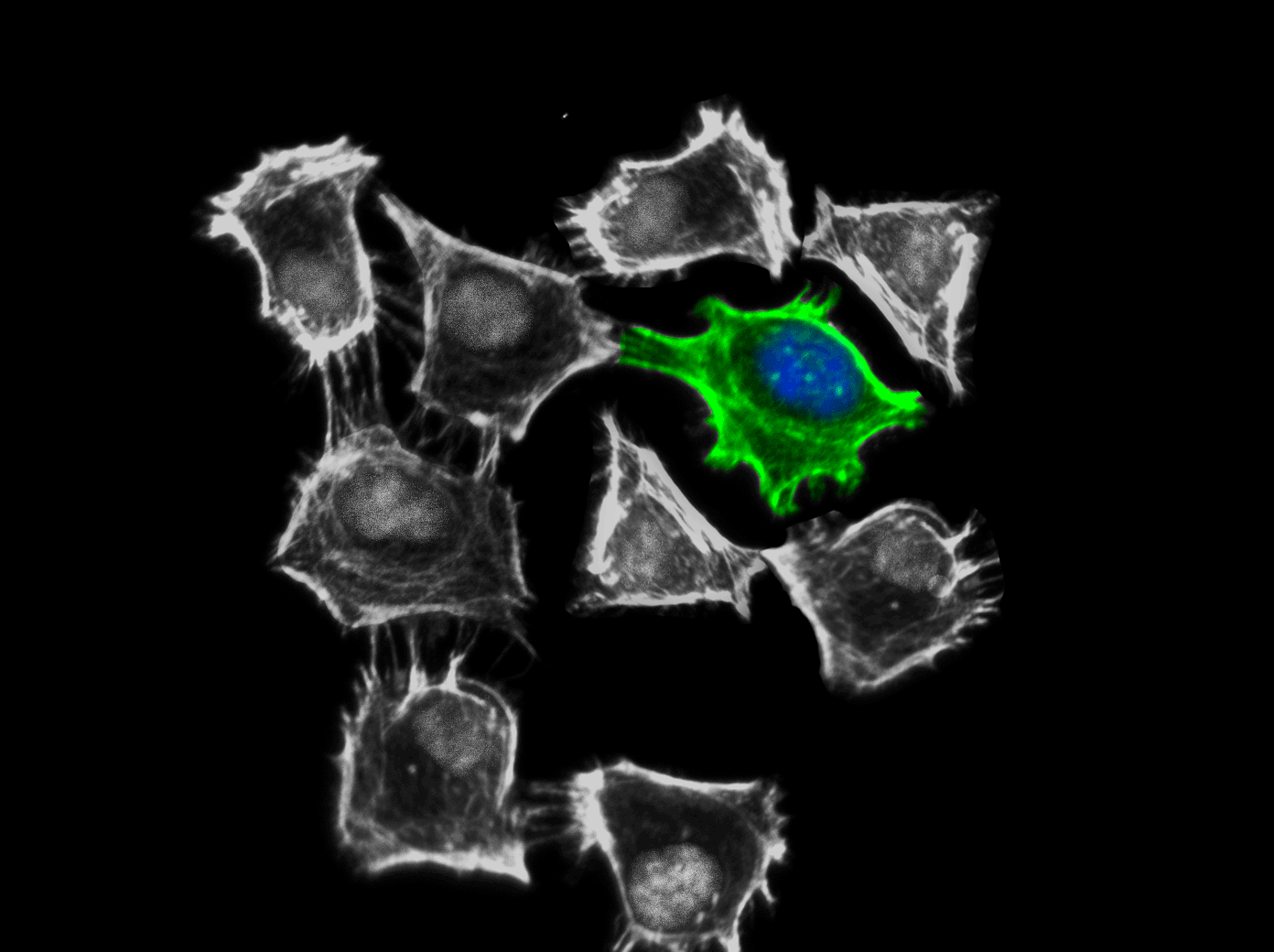
For more information about Cdc42, please see here:
Abdul-Manan N. et al. 1999. Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein. Nature. 399, 379-383.
Abraham M.T. et al. 2001. Motility-related proteins as markers for head and neck squamous cell cancer. Laryngoscope. 111, 1285-1289.
Benitah S.A. et al. 2004. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta. 1705, 121-132.
Bishop A.L. and Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348, 241-255.
Boettner B. and Van Aelst L. 2002. The role of Rho GTPases in disease development. Gene. 286, 155-174.
Cappello S. et al. 2006. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat. Neurosci. 9, 1099-1107.
Cerione R.A. 2004. Cdc42: new roads to travel. Trends Cell Biol. 14, 127-132.
Fritz G. et al. 2002. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer. 87, 635-644.
Fritz G. et al. 1999. Rho GTPases are over-expressed in human tumors. Int. J. Cancer. 81, 682-687.
Gomez del Pulgar T. et al. 2005. In Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 27, 602-613.
Gupton S.L. and Gertler F.B. 2007. Filopodia: the fingers that do the walking. Sci. STKE. 2007, re5.
Heasman S.J. and Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690-701.
Sinha S. and Yang W. 2008. Cellular signaling for activation of Rho GTPase Cdc42. Cell Signal. 20, 1927-1934.
Symons M. and Settleman J. 2000. Rho family GTPases: more than simple switches. Trends Cell Biol. 10, 415-419.
Vega F.M. and Ridley A.J. 2008. Rho GTPases in cancer cell biology. FEBS Lett. 582, 2093-2101.
Rac/Cdc42 Activator II (Cat. # CN02)
Rho/Rac/Cdc42 Activator I (Cat. # CN04)
Acti-stain 488 phalloidin (Cat. # PHDG1-A)
Acti-stain 555 phalloidin (Cat. # PHDH1-A)
Acti-stain 670 phalloidin (Cat. # PHDN1-A)
Rhodamine Phalloidin (Cat. # PHDR1)
SiR-Actin Kit 50 nmol SiR-Actin and 1 umol verapamil (Cat. # CY-SC001)
Anti-Cdc42: mouse Mab (Cat. # ACD03)
Cdc42 G-LISA Activation Assay (Colorimetric format) - 96 assays (Cat. # BK127)
Cdc42 G-LISA Activation Assay (Colorimetric format) - 24 assays (Cat. # BK127-S)
Cdc42 Pull-down Activation Assay Biochem Kit (bead pull-down format) - 50 Assays (Cat. # BK034)
Cdc42 Pull-down Activation Assay Biochem Kit (bead pull-down format) - 20 Assays (Cat. # BK034-S)
Cdc42 protein: GST tagged: human dominant negative (Cat. # C17G01)
Cdc42 protein: GST tagged: human wild type (Cat. # CDG01)
Cdc42 protein: His tagged: human constitutively active (Cat. # C6101)